For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTTASQAWAQAMAFAHRADPYPFFAELRKTPVARVADDLYVVTGYYELLALAHDPRVSSDFRRSPLSSDR APAEPADDVGAYGSEPSLIVTDPPSHDRARRQVSRHFAPPHSPDLIPSMEPAVVALCNDLLDTAKANGRT RIDVVDDYAYPVPVMVICKILGVPLEDEPRFHAWIFDMLAGVDLGPDATTDEGQGRAARARDAQAALRDY LAGLIEGYRGAGGDGLLSALVNDASGPDGAMPPGQALANAVLLLIAGHDSTVNTISNCVLTFLRNPGTVE LLRDRPELIPRAIEEVQRLQSAVQFFPSRSATADIEVGGTVIPEGAAVHLMYGAANRDPRRFADPDTFDI FRPDNEHFGWGSGIHTCVGGPLARLEVNLALEIFLRRVHRPRLVSDPPPYRQSQIFRGPRHVLVDVEAIG D
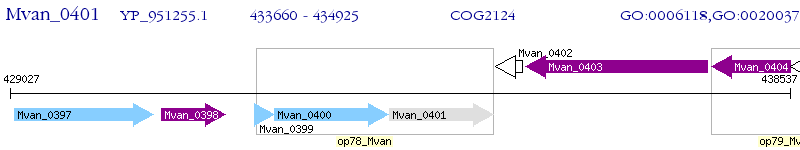
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0401 | - | - | 100% (421) | cytochrome P450 |
| M. vanbaalenii PYR-1 | Mvan_5579 | - | 8e-53 | 35.22% (423) | cytochrome P450 |
| M. vanbaalenii PYR-1 | Mvan_5525 | - | 7e-41 | 35.54% (332) | cytochrome P450 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1912c | cyp140 | 2e-38 | 31.83% (399) | cytochrome p450 140 CYP140 |
| M. gilvum PYR-GCK | Mflv_1229 | - | 4e-57 | 37.77% (413) | cytochrome P450 |
| M. tuberculosis H37Rv | Rv1880c | cyp140 | 2e-38 | 31.83% (399) | cytochrome p450 140 CYP140 |
| M. leprae Br4923 | MLBr_02088 | - | 3e-52 | 34.96% (409) | putative cytochrome p450 |
| M. abscessus ATCC 19977 | MAB_0276 | - | 8e-49 | 33.90% (413) | cytochrome P450 |
| M. marinum M | MMAR_2930 | cyp147G1 | 1e-169 | 68.74% (419) | cytochrome P450 147G1 Cyp147G1 |
| M. avium 104 | MAV_0358 | - | 7e-61 | 36.94% (425) | cytochrome P450 |
| M. smegmatis MC2 155 | MSMEG_6312 | - | 2e-52 | 35.95% (395) | cytochrome P450 107B1 |
| M. thermoresistible (build 8) | TH_2944 | - | 1e-41 | 31.89% (417) | PUTATIVE cytochrome P450 |
| M. ulcerans Agy99 | MUL_2979 | cyp140A5 | 2e-36 | 30.75% (400) | cytochrome P450 140A5 Cyp140A5 |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_0401|M.vanbaalenii_PYR-1 ------------------------------MTTASQAWAQAMAFAHRADP
MMAR_2930|M.marinum_M -------------------------------MNAETAWAEAMKFENRPNP
Mflv_1229|M.gilvum_PYR-GCK ---------------------------MTTAAEPQSLLLQMLDPANRADP
MSMEG_6312|M.smegmatis_MC2_155 -------------------MHNGWMSTAATAQEAQGLLLQLLDPATRADP
MAV_0358|M.avium_104 -----------------------MTTAPTESAESQGLLLQLLDPANRADP
MLBr_02088|M.leprae_Br4923 MRTCVPTRTCVYAFIEYLSHNRPMGTNPPSLVEAQMLLLRLIDPGTRADP
MAB_0276|M.abscessus_ATCC_1997 ---------------------------MPITTDPGTLLLQVLDPANRANP
Mb1912c|M.bovis_AF2122/97 ---------MKDKLHWLAMHGVIRGIAAIGIRRGDLQARLIADPAVATDP
Rv1880c|M.tuberculosis_H37Rv ---------MKDKLHWLAMHGVIRGIAAIGIRRGDLQARLIADPAVATDP
MUL_2979|M.ulcerans_Agy99 ---------MKDKLHWFAMHGVIRGMANIGLRRGELEARLIADPAVAANP
TH_2944|M.thermoresistible__bu -------------------------------------MTAAKPAGYLRDP
:*
Mvan_0401|M.vanbaalenii_PYR-1 YPFFAELRKTP-VARVADDLYVVTGYYELLALAHDPRVSSDFRRSP--LS
MMAR_2930|M.marinum_M YPYFDELRKTP-VAKVAEKTYVVTGYRELLALAHDPRISSDITRSPSGFG
Mflv_1229|M.gilvum_PYR-GCK YPLYARIRAQ---GPVQLPANNLTVFSTYAA---CDEILRHPHASSDRLK
MSMEG_6312|M.smegmatis_MC2_155 YPIYDRIRRG---GPLALPEANLAVFSSFSD---CDDVLRHPSSCSDRTK
MAV_0358|M.avium_104 YRLYAQFRER---GALQLPEANLAVFLSYRD---CDEVLRHPSSSSDNVN
MLBr_02088|M.leprae_Br4923 FPVYRALIDY---GPMQLPGMPLTVFSSFSD---CDEALRHPLSASDRLK
MAB_0276|M.abscessus_ATCC_1997 YPVYGQLVEQTQAGPVHPPGMDVSVLATFAD---CNAVLRHPQASSDRRK
Mb1912c|M.bovis_AF2122/97 VPFYDEVRSHG--ALVRNRANYLTVDHRLAHDLLRSDDFRVVSFGENLPP
Rv1880c|M.tuberculosis_H37Rv VPFYDEVRSHG--ALVRNRANYLTVDHRLAHDLLRSDDFRVVSFGENLPP
MUL_2979|M.ulcerans_Agy99 GPFHDELRRHG--RMVRTRVNYLTVDYQLAHDVLRSDDFHTISFGENLPG
TH_2944|M.thermoresistible__bu YPFFAERRRGS--GVFHGTVIDYSRTPESLR-PTDGYAAMSFDAVNRVLR
. . :
Mvan_0401|M.vanbaalenii_PYR-1 SDRAPAEP-ADDVGAYGSEPSLIVTDPPSHDRARRQVSRHFAPPHSPDLI
MMAR_2930|M.marinum_M GEAPQPEPGSEHVQAYGQDASIIVSDPPDHDRARRQVMRHFAPPHSPDLI
Mflv_1229|M.gilvum_PYR-GCK STLAQRAIADGAQPRPFGPPGFLFLDPPDHTRLRRLVSKAFVP----KVV
MSMEG_6312|M.smegmatis_MC2_155 STIFQRQLAAETQPRPQGPASFLFLDPPDHTRLRGLVSKAFAP----RVI
MAV_0358|M.avium_104 STVAKRQAAAGTAPVRQGPPGFLFLDPPDHTRLRRLVSKAFAP----RVV
MLBr_02088|M.leprae_Br4923 ATLAQQAIAAGAEPRPFYASSFMFLDPPDHTRLRKLVSKAFAP----KVV
MAB_0276|M.abscessus_ATCC_1997 SNIVQRQLAQ--NPNLIARPSFLGLDAPDHTRLRKLASKAFAP----RVI
Mb1912c|M.bovis_AF2122/97 PLRWLERRTRGDQLHPLREPSLLAVEPPDHTRYRKTVSAVFTS----RAV
Rv1880c|M.tuberculosis_H37Rv PLRWLERRTRGDQLHPLREPSLLAVEPPDHTRYRKTVSAVFTS----RAV
MUL_2979|M.ulcerans_Agy99 PLRWLERRTRDEQLHPLRPPSLLAVEPPDHTRYRKTVSAVFTS----RAV
TH_2944|M.thermoresistible__bu DGRVFSSALYDTTIGLFTGPTLLAMEGGTHRAHRNLISAAFKT----KAL
. :: : * * * . :
Mvan_0401|M.vanbaalenii_PYR-1 PSMEPAVVA-LCNDLLDTAKANGRTRIDVVDDYAYPVPVMVICKILGVPL
MMAR_2930|M.marinum_M PSMEPFVVR-LANDLLNQASARGSTRLDVVEDFAYPIPVAVICKILGVPI
Mflv_1229|M.gilvum_PYR-GCK KALEPEIVA-LVDALLA-DAGG---TFDAIEGLAYPLPVAVICRLLGVPL
MSMEG_6312|M.smegmatis_MC2_155 KRLEPEITA-LVDQLLD-AVDGP--EFNLIDNLAYPLPVAVICRLLGVPI
MAV_0358|M.avium_104 AALEPDIRS-LVDGLLDRAADRG--ELEIVEDFAYPLPVAVICRLLGVPL
MLBr_02088|M.leprae_Br4923 QALEGDIAA-LVDSLLDKGAAAG--QFDVIADLAFPLAVAVICRLLGVPY
MAB_0276|M.abscessus_ATCC_1997 NAMAEDIQT-FVDKMLDDIALRG--TFNLVTEFAYPLPVAVICRMLGVPI
Mb1912c|M.bovis_AF2122/97 SALRDLVEQ-TAINLLDRFAEQPG-IVDVVGRYCSQLPIVVISEILGVPE
Rv1880c|M.tuberculosis_H37Rv SALRDLVEQ-TAINLLDRFAEQPG-IVDVVGRYCSQLPIVVISEILGVPE
MUL_2979|M.ulcerans_Agy99 AALRDRVEQ-TANSLLDQLAQRPG-VVDVVGQYCSQLPIMVISEILGVPE
TH_2944|M.thermoresistible__bu ARWEPSIVRPIVNALIDEFIDTG--TADLVRDFTFEFPTRVISRMLGLPE
: :: : : .. **..:**:*
Mvan_0401|M.vanbaalenii_PYR-1 EDEPRFHAWIFDMLAGVDLGPDATTDEGQGRAARARDAQAALRDYLAGLI
MMAR_2930|M.marinum_M EDEPKFHAWIFDFMAGTDLGPEGDTEEGQVLAEKGRVSTAALTDYLGDLV
Mflv_1229|M.gilvum_PYR-GCK ADEPEFSAASALLAQSLDPFATVTGSAGDGFEER-MEAARWLRGYLRELI
MSMEG_6312|M.smegmatis_MC2_155 EDEPKFSRASALLAAALDPFLALTGETSDLFDEQ-MKAGMWLRDYLRALI
MAV_0358|M.avium_104 DDEPQFSRASALLAQALDPFSTITGVPAEVASER-QQAGTWLRDYFHQLI
MLBr_02088|M.leprae_Br4923 EDAPEFGRVSALLVQSVDPFITITGEPPEATEER-LRAGVWLRDYLEQLV
MAB_0276|M.abscessus_ATCC_1997 EDEPEFGRASALLGQGLDPVYALTGQSPANMDER-FQAARWMWDYFIDLI
Mb1912c|M.bovis_AF2122/97 HDRPRVLEFGELAAPSLD-----IGIPWRQYLRV-QQGIRGFDCWLEGHL
Rv1880c|M.tuberculosis_H37Rv HDRPRVLEFGELAAPSLD-----IGIPWRQYLRV-QQGIRGFDCWLEGHL
MUL_2979|M.ulcerans_Agy99 QDRGRVLEYGELAAPSLD-----IGVTYRQYRRI-QQGITGFNSWLTAHL
TH_2944|M.thermoresistible__bu EDLPTFRRWAVQLISYTVD------------YQRAFEASAALKDYFLEQM
* . . : :: :
Mvan_0401|M.vanbaalenii_PYR-1 EGYRGAGGDGLLSALVNDASGPDGA--MPPGQALANAVLLLIAGHDSTVN
MMAR_2930|M.marinum_M RSYAKTPGEGLLSKLLHDD-GPDGP--MSVPETTANALLLLVAGHDSTVN
Mflv_1229|M.gilvum_PYR-GCK SQRRRDPGEDLMSGLIQVEESGD---QLTEDEIVATCNLLLVAGHETTVN
MSMEG_6312|M.smegmatis_MC2_155 DERRRTPGEDLMSGLVAVEESGD---QLTEDEIIATCNLLLIAGHETTVN
MAV_0358|M.avium_104 EARRSRPGDDLLSGLIAVEESGD---QLTEEEIVSTCNLLLIAGHETTVN
MLBr_02088|M.leprae_Br4923 KCRRGTPGEDLISRLIELDESGD---QLTEEEIIATCGLLLVAGHETTVN
MAB_0276|M.abscessus_ATCC_1997 ASRRRNLGDDLLSALIQVEEAGD---QLTEEEIISTCTLLLIAGHETTVN
Mb1912c|M.bovis_AF2122/97 QQLRHAPGDDLMSQLIQIAESGDNETQLDETELRAIAGLVLVAGFETTVN
Rv1880c|M.tuberculosis_H37Rv QQLRHAPGDDLMSQLIQIAESGDNETQLDETELRAIAGLVLVAGFETTVN
MUL_2979|M.ulcerans_Agy99 EQLRRNPGDDLMSQLIRVAESGDAGTYLDKSELRAIAGLVLVAGFETTVN
TH_2944|M.thermoresistible__bu ESRRSKPTDDMIGDLVTAEIDGE---KLSDEAIYSFLRLLLPAGLETTYR
:.::. *: : : : *:* ** ::* .
Mvan_0401|M.vanbaalenii_PYR-1 TISNCVLTFLRNPGTVELLRDRPELIPRAIEEVQRLQSAVQFFPSRSATA
MMAR_2930|M.marinum_M TITNCVMTLLRNPGSWDLVRQRPELIPRTIEEVQRLQSAVQFFPSRSATD
Mflv_1229|M.gilvum_PYR-GCK LIANAILALLSNDGQWSALAADPARVGAVVEETLRYDPPVQLVG-RIAAE
MSMEG_6312|M.smegmatis_MC2_155 LIANAALAMLRTPGQWAALAADGSRASAVIEETMRYDPPVQLVS-RYAGD
MAV_0358|M.avium_104 LIGNAVLAMLRDPGQWAALGADPGRAPAIVEETLRYDPPVQLAG-QIALD
MLBr_02088|M.leprae_Br4923 LIANAVLAMLRNPSQWKALSSNPQRAPLVVEETLRYDPAIHLIG-RVAAK
MAB_0276|M.abscessus_ATCC_1997 LIANASLAMLRHPEQWKLLGQNADRAPLVVEETLRYDPPVHMVA-RVADG
Mb1912c|M.bovis_AF2122/97 LLGNGIRMLLDTPEHLATLRQHPELWPNTVEEILRLDSPVQLTA-RVACR
Rv1880c|M.tuberculosis_H37Rv LLGNGIRMLLDTPEHLATLRQHPELWPNTVEEILRLDSPVQLTA-RVACR
MUL_2979|M.ulcerans_Agy99 LLGNGIRMLLDAPEQLETLRQRPELWPNAVEEILRLDSPVQLTA-RVART
TH_2944|M.thermoresistible__bu SSGNLIYLLLTHPEQFAAVAADPGLIPQAIEEGLRHETPLTTVQ-RLTTE
* :* : :** * :..: : :
Mvan_0401|M.vanbaalenii_PYR-1 DIEVGGTVIPEGAAVHLMYGAANRDPRRFADPDTFDIFRPDN-EHFGWGS
MMAR_2930|M.marinum_M EIEIGGTVIPAGSAVHLIYAAANRDPRRFDNPNRFDPLREDN-EHFGWGS
Mflv_1229|M.gilvum_PYR-GCK DMTVGTTTVPKGDNMLLLTAAAHRDPDAVDRPDEFDPDRESF-RHLGFGK
MSMEG_6312|M.smegmatis_MC2_155 DLTIGTHTVPKGDTMLLLLAAAHRDPTIVGAPDRFDPDRAQI-RHLGFGK
MAV_0358|M.avium_104 DMVIGGVEVPAGDVMMLLLAAANRDPAEFDRPDTFDPDRKSL-RHLGFGR
MLBr_02088|M.leprae_Br4923 DMTIGQTTLTEGDTMVLLLAAANRDPAVYSRPDEFDPDRPSS-RHLAFAV
MAB_0276|M.abscessus_ATCC_1997 EMAIRDFILPDGEWVLLMLAAAQRDPDVGPDLDVFNPDRSEI-KHLAFGH
Mb1912c|M.bovis_AF2122/97 DVEVAGVRIKRGEVVVIYLAAANRDPAVFPDPHRFDIERPNAGRHLAFST
Rv1880c|M.tuberculosis_H37Rv DVEVAGVRIKRGEVVVIYLAAANRDPAVFPDPHRFDIERPNAGRHLAFST
MUL_2979|M.ulcerans_Agy99 DVEIASWPINRGEMVVAYLAGANRDPAVFPNPHRFDVERANAGRHLAFST
TH_2944|M.thermoresistible__bu DTELCGAAIPAGAVVDTCIGSANRDETRWDRPEEFDIFRDRV-PHLSFAA
: : : * : ..*:** . *: * *:.:.
Mvan_0401|M.vanbaalenii_PYR-1 GIHTCVGGPLARLEVNLALEIFLRRVHRPRLVSDPPPYRQSQIFRGPRHV
MMAR_2930|M.marinum_M GIHTCMGGPLARLEVNLAVEIFLRRVQSPKLVVDPPPYRHNQIFRGPRHL
Mflv_1229|M.gilvum_PYR-GCK GPHFCIGAPLARLEAAVALTALTSRFPGAQLA-GEPSYKQNVTLRGMSEL
MSMEG_6312|M.smegmatis_MC2_155 GAHFCLGAPLARLEATVALPALAARFPEARLS-GEPEYKRNLTLRGMSTL
MAV_0358|M.avium_104 GVHYCLGAPLARLEAGVALSAVTARFPRARLD-GEPQYKTNVTLRGLSRL
MLBr_02088|M.leprae_Br4923 GSHFCLGAALARLEATVTLSAISARFPQVQLA-GELVYKPNVAMRGMSAL
MAB_0276|M.abscessus_ATCC_1997 GPHFCLGAPLARLEARLALSSITARFPGARLM-DEPTYKPNVTLRGLAEL
Mb1912c|M.bovis_AF2122/97 GRHFCLGAALARAEGEVGLRTFFDRFPDVRAA-GAGSRRDTRVLRGWSTL
Rv1880c|M.tuberculosis_H37Rv GRHFCLGAALARAEGEVGLRTFFDRFPDVRAA-GAGSRRDTRVLRGWSTL
MUL_2979|M.ulcerans_Agy99 GRHFCLGAALARAEGEVGLRTFFERFPDVRAA-GAGSRRDTRVLRGWSTL
TH_2944|M.thermoresistible__bu GEHTCMGLHLARMETRVALETLLSRVHDLRLVTDGDPHIFGQPFRSPTAI
* * *:* *** * : : . *. : . :*. :
Mvan_0401|M.vanbaalenii_PYR-1 LVDVEAIGD----
MMAR_2930|M.marinum_M WVDFAAITE----
Mflv_1229|M.gilvum_PYR-GCK PITL---------
MSMEG_6312|M.smegmatis_MC2_155 SIAV---------
MAV_0358|M.avium_104 VVAV---------
MLBr_02088|M.leprae_Br4923 PVQV---------
MAB_0276|M.abscessus_ATCC_1997 PVSIA--------
Mb1912c|M.bovis_AF2122/97 PVTLGPARSMVSP
Rv1880c|M.tuberculosis_H37Rv PVTLGPARSMVSP
MUL_2979|M.ulcerans_Agy99 PVTLGPARALAAS
TH_2944|M.thermoresistible__bu PVTFDPVR-----
: .