For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTTAPTESAESQGLLLQLLDPANRADPYRLYAQFRERGALQLPEANLAVFLSYRDCDEVLRHPSSSSDNV NSTVAKRQAAAGTAPVRQGPPGFLFLDPPDHTRLRRLVSKAFAPRVVAALEPDIRSLVDGLLDRAADRGE LEIVEDFAYPLPVAVICRLLGVPLDDEPQFSRASALLAQALDPFSTITGVPAEVASERQQAGTWLRDYFH QLIEARRSRPGDDLLSGLIAVEESGDQLTEEEIVSTCNLLLIAGHETTVNLIGNAVLAMLRDPGQWAALG ADPGRAPAIVEETLRYDPPVQLAGQIALDDMVIGGVEVPAGDVMMLLLAAANRDPAEFDRPDTFDPDRKS LRHLGFGRGVHYCLGAPLARLEAGVALSAVTARFPRARLDGEPQYKTNVTLRGLSRLVVAV
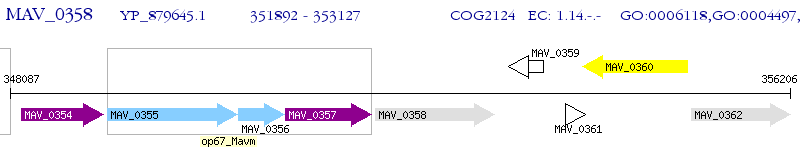
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_0358 | - | - | 100% (411) | cytochrome P450 |
| M. avium 104 | MAV_1274 | - | 1e-58 | 39.10% (376) | cytochrome P450 superfamily protein |
| M. avium 104 | MAV_2822 | - | 6e-57 | 37.14% (412) | P450 heme-thiolate protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1912c | cyp140 | 1e-61 | 38.07% (415) | cytochrome p450 140 CYP140 |
| M. gilvum PYR-GCK | Mflv_1229 | - | 1e-155 | 66.50% (406) | cytochrome P450 |
| M. tuberculosis H37Rv | Rv1880c | cyp140 | 1e-61 | 38.07% (415) | cytochrome p450 140 CYP140 |
| M. leprae Br4923 | MLBr_02088 | - | 1e-142 | 60.34% (411) | putative cytochrome p450 |
| M. abscessus ATCC 19977 | MAB_0276 | - | 1e-125 | 57.61% (401) | cytochrome P450 |
| M. marinum M | MMAR_5268 | cyp164A3 | 1e-169 | 71.70% (417) | cytochrome P450 164A3 Cyp164A3 |
| M. smegmatis MC2 155 | MSMEG_6312 | - | 1e-158 | 67.64% (411) | cytochrome P450 107B1 |
| M. thermoresistible (build 8) | TH_1650 | - | 3e-82 | 61.20% (250) | cytochrome P450 107B1 |
| M. ulcerans Agy99 | MUL_2979 | cyp140A5 | 3e-58 | 36.95% (406) | cytochrome P450 140A5 Cyp140A5 |
| M. vanbaalenii PYR-1 | Mvan_5579 | - | 1e-150 | 64.75% (417) | cytochrome P450 |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_0358|M.avium_104 ------------------------MTTAP------TESAESQGLLLQLLD
MMAR_5268|M.marinum_M MLRGPVTAATHFDHTFFMRRHNVSMTTRPIEPIEFTEQVEPQAFLFQLLD
Mflv_1229|M.gilvum_PYR-GCK ----------------------------------MTTAAEPQSLLLQMLD
Mvan_5579|M.vanbaalenii_PYR-1 ----------------------------------MTTAAEPQSLLLQMLA
MSMEG_6312|M.smegmatis_MC2_155 ------------------------MHNGWMS--TAATAQEAQGLLLQLLD
MLBr_02088|M.leprae_Br4923 -------MRTCVPTRTCVYAFIEYLSHNRPMGTNPPSLVEAQMLLLRLID
MAB_0276|M.abscessus_ATCC_1997 ----------------------------------MPITTDPGTLLLQVLD
TH_1650|M.thermoresistible__bu ----------------------------------MTVEADPQILLMQMLD
Mb1912c|M.bovis_AF2122/97 ----------------MKDKLHWLAMHGVIRGIAAIGIRRGDLQARLIAD
Rv1880c|M.tuberculosis_H37Rv ----------------MKDKLHWLAMHGVIRGIAAIGIRRGDLQARLIAD
MUL_2979|M.ulcerans_Agy99 ----------------MKDKLHWFAMHGVIRGMANIGLRRGELEARLIAD
:
MAV_0358|M.avium_104 PANRADPYRLYAQFRER---GALQLPEANLAVFLSYRD---CDEVLRHPS
MMAR_5268|M.marinum_M PAIRADPYPLCARLRDH---GPLHLPEANFVLFSSFGD---CDEVLRHPS
Mflv_1229|M.gilvum_PYR-GCK PANRADPYPLYARIRAQ---GPVQLPANNLTVFSTYAA---CDEILRHPH
Mvan_5579|M.vanbaalenii_PYR-1 PEVRADPYPVYAQILAH---GPMQLPGNNLTVFSSYAD---CDEVLRHPD
MSMEG_6312|M.smegmatis_MC2_155 PATRADPYPIYDRIRRG---GPLALPEANLAVFSSFSD---CDDVLRHPS
MLBr_02088|M.leprae_Br4923 PGTRADPFPVYRALIDY---GPMQLPGMPLTVFSSFSD---CDEALRHPL
MAB_0276|M.abscessus_ATCC_1997 PANRANPYPVYGQLVEQTQAGPVHPPGMDVSVLATFAD---CNAVLRHPQ
TH_1650|M.thermoresistible__bu PANRSDPYPVYRRIRER---GPVQPAGGNVTVFSSYAD---CDAVLRHPD
Mb1912c|M.bovis_AF2122/97 PAVATDPVPFYDEVRSHG--ALVRNRANYLTVDHRLAHDLLRSDDFRVVS
Rv1880c|M.tuberculosis_H37Rv PAVATDPVPFYDEVRSHG--ALVRNRANYLTVDHRLAHDLLRSDDFRVVS
MUL_2979|M.ulcerans_Agy99 PAVAANPGPFHDELRRHG--RMVRTRVNYLTVDYQLAHDVLRSDDFHTIS
* ::* . . : . : . ::
MAV_0358|M.avium_104 SSSDNVNSTVAKRQAAAGTAPVRQGPPGFLFLDPPDHTRLRRLVSKAFAP
MMAR_5268|M.marinum_M SCSDQTKSTVAQRRVEEEPALGRDFPPGFLFLDPPDHTRLRKLVSKAFAP
Mflv_1229|M.gilvum_PYR-GCK ASSDRLKSTLAQRAIADGAQPRPFGPPGFLFLDPPDHTRLRRLVSKAFVP
Mvan_5579|M.vanbaalenii_PYR-1 ASSDRLKSTAAQRAIADGAEARPFGPPGFLFLDPPDHTRLRWLVSKAFVP
MSMEG_6312|M.smegmatis_MC2_155 SCSDRTKSTIFQRQLAAETQPRPQGPASFLFLDPPDHTRLRGLVSKAFAP
MLBr_02088|M.leprae_Br4923 SASDRLKATLAQQAIAAGAEPRPFYASSFMFLDPPDHTRLRKLVSKAFAP
MAB_0276|M.abscessus_ATCC_1997 ASSDRRKSNIVQRQLAQN--PNLIARPSFLGLDAPDHTRLRKLASKAFAP
TH_1650|M.thermoresistible__bu SCSDGLKSTITRRQLAEGKDVRPLGPPGFLFLDPPDHTRLRGLVSKAFAP
Mb1912c|M.bovis_AF2122/97 FGENLPPPLRWLERRTRGDQLHPLREPSLLAVEPPDHTRYRKTVSAVFTS
Rv1880c|M.tuberculosis_H37Rv FGENLPPPLRWLERRTRGDQLHPLREPSLLAVEPPDHTRYRKTVSAVFTS
MUL_2979|M.ulcerans_Agy99 FGENLPGPLRWLERRTRDEQLHPLRPPSLLAVEPPDHTRYRKTVSAVFTS
.: . . ..:: ::.***** * .* .*..
MAV_0358|M.avium_104 RVVAALEPDIRSLVDGLLDRAADRGE-LEIVEDFAYPLPVAVICRLLGVP
MMAR_5268|M.marinum_M KVVNALRPEISSLVDGLLDRIAEQGR-FDVVSDFAYPLPVAVICRLLGVP
Mflv_1229|M.gilvum_PYR-GCK KVVKALEPEIVALVDALL--ADAGGT-FDAIEGLAYPLPVAVICRLLGVP
Mvan_5579|M.vanbaalenii_PYR-1 KVVKALEPDIVTMVDGLLREAEAGAT-FEAIGGLAYPLPVAVICRLLGVP
MSMEG_6312|M.smegmatis_MC2_155 RVIKRLEPEITALVDQLLD-AVDGPE-FNLIDNLAYPLPVAVICRLLGVP
MLBr_02088|M.leprae_Br4923 KVVQALEGDIAALVDSLLDKGAAAGQ-FDVIADLAFPLAVAVICRLLGVP
MAB_0276|M.abscessus_ATCC_1997 RVINAMAEDIQTFVDKMLDDIALRGT-FNLVTEFAYPLPVAVICRMLGVP
TH_1650|M.thermoresistible__bu KVVRALEADIAAMVDELLAAAEPGGEPFDVISGLAYPLPVTVICRLLGVP
Mb1912c|M.bovis_AF2122/97 RAVSALRDLVEQTAINLLDRFAEQPGIVDVVGRYCSQLPIVVISEILGVP
Rv1880c|M.tuberculosis_H37Rv RAVSALRDLVEQTAINLLDRFAEQPGIVDVVGRYCSQLPIVVISEILGVP
MUL_2979|M.ulcerans_Agy99 RAVAALRDRVEQTANSLLDQLAQRPGVVDVVGQYCSQLPIMVISEILGVP
:.: : : . :* .: : . *.: **..:****
MAV_0358|M.avium_104 LDDEPQFSRASALLAQALDPFSTITGVPAEVASERQQAGTWLRDYFHQLI
MMAR_5268|M.marinum_M LEDEPQFSQASALLAQALDPFFAFTGQNAENLNDSLEAGQWLRGYLRDLI
Mflv_1229|M.gilvum_PYR-GCK LADEPEFSAASALLAQSLDPFATVTGSAGDGFEERMEAARWLRGYLRELI
Mvan_5579|M.vanbaalenii_PYR-1 LEDEPQFSAASALLAQSLDPFISVTGSAGSGFEERMQAARWLRGYLRELI
MSMEG_6312|M.smegmatis_MC2_155 IEDEPKFSRASALLAAALDPFLALTGETSDLFDEQMKAGMWLRDYLRALI
MLBr_02088|M.leprae_Br4923 YEDAPEFGRVSALLVQSVDPFITITGEPPEATEERLRAGVWLRDYLEQLV
MAB_0276|M.abscessus_ATCC_1997 IEDEPEFGRASALLGQGLDPVYALTGQSPANMDERFQAARWMWDYFIDLI
TH_1650|M.thermoresistible__bu IEDEPRFRDASALLAQGLDPFLTVTGQAADGHEQRVAAGQWLRGYLGELL
Mb1912c|M.bovis_AF2122/97 EHDRPRVLEFGELAAPSLD-----IGIPWRQYLRVQQGIRGFDCWLEGHL
Rv1880c|M.tuberculosis_H37Rv EHDRPRVLEFGELAAPSLD-----IGIPWRQYLRVQQGIRGFDCWLEGHL
MUL_2979|M.ulcerans_Agy99 EQDRGRVLEYGELAAPSLD-----IGVTYRQYRRIQQGITGFNSWLTAHL
* .. . * .:* * . : :: :
MAV_0358|M.avium_104 EARRSRPGDDLLSGLIAVEESGD---QLTEEEIVSTCNLLLIAGHETTVN
MMAR_5268|M.marinum_M ALRRSQPGEDLMSGLVAVEESGD---QLTEDEIVSTCVLLLVAGHETTVN
Mflv_1229|M.gilvum_PYR-GCK SQRRRDPGEDLMSGLIQVEESGD---QLTEDEIVATCNLLLVAGHETTVN
Mvan_5579|M.vanbaalenii_PYR-1 ARRRRDPGEDLMSGLIHVEESGD---QLTEDEIVATCNLLLVAGHETTVN
MSMEG_6312|M.smegmatis_MC2_155 DERRRTPGEDLMSGLVAVEESGD---QLTEDEIIATCNLLLIAGHETTVN
MLBr_02088|M.leprae_Br4923 KCRRGTPGEDLISRLIELDESGD---QLTEEEIIATCGLLLVAGHETTVN
MAB_0276|M.abscessus_ATCC_1997 ASRRRNLGDDLLSALIQVEEAGD---QLTEEEIISTCTLLLIAGHETTVN
TH_1650|M.thermoresistible__bu EARRADPRDDLMSALIEVVESGD---QLSTEEIIATCNLLLVAGXXX---
Mb1912c|M.bovis_AF2122/97 QQLRHAPGDDLMSQLIQIAESGDNETQLDETELRAIAGLVLVAGFETTVN
Rv1880c|M.tuberculosis_H37Rv QQLRHAPGDDLMSQLIQIAESGDNETQLDETELRAIAGLVLVAGFETTVN
MUL_2979|M.ulcerans_Agy99 EQLRRNPGDDLMSQLIRVAESGDAGTYLDKSELRAIAGLVLVAGFETTVN
* :**:* *: : *:** * *: : . *:*:**
MAV_0358|M.avium_104 LIGNAVLAMLRDPGQWAALGADPGRAPAIVEETLRYDPPVQLAGQIALDD
MMAR_5268|M.marinum_M LIGNAILAMLRDRRQWAQLGADPDRAAAVVEETMRYDPPVQLIGRIAGAD
Mflv_1229|M.gilvum_PYR-GCK LIANAILALLSNDGQWSALAADPARVGAVVEETLRYDPPVQLVGRIAAED
Mvan_5579|M.vanbaalenii_PYR-1 LIANAILALLRNDGQWAALAADPGRAAAVIEETLRHDPPVQLVGRIAGQD
MSMEG_6312|M.smegmatis_MC2_155 LIANAALAMLRTPGQWAALAADGSRASAVIEETMRYDPPVQLVSRYAGDD
MLBr_02088|M.leprae_Br4923 LIANAVLAMLRNPSQWKALSSNPQRAPLVVEETLRYDPAIHLIGRVAAKD
MAB_0276|M.abscessus_ATCC_1997 LIANASLAMLRHPEQWKLLGQNADRAPLVVEETLRYDPPVHMVARVADGE
TH_1650|M.thermoresistible__bu --------------AGGGAGPGPRRG-------QRTGGADHCADRCAAP-
Mb1912c|M.bovis_AF2122/97 LLGNGIRMLLDTPEHLATLRQHPELWPNTVEEILRLDSPVQLTARVACRD
Rv1880c|M.tuberculosis_H37Rv LLGNGIRMLLDTPEHLATLRQHPELWPNTVEEILRLDSPVQLTARVACRD
MUL_2979|M.ulcerans_Agy99 LLGNGIRMLLDAPEQLETLRQRPELWPNAVEEILRLDSPVQLTARVARTD
* . . : : *
MAV_0358|M.avium_104 MVIG-----------GVEVPAGDVMMLLLAAANRDPAEFDRPDTFDPDRK
MMAR_5268|M.marinum_M MVIG-----------DVEVAKGDVMMMLVAAAHRDPAEYDRPEEFDPDRG
Mflv_1229|M.gilvum_PYR-GCK MTVG-----------TTTVPKGDNMLLLTAAAHRDPDAVDRPDEFDPDRE
Mvan_5579|M.vanbaalenii_PYR-1 MTIGGERSDGGNVAGGVTVPKGDNILLLTAAAHRDPDHVDRPDEFDPDRP
MSMEG_6312|M.smegmatis_MC2_155 LTIG-----------THTVPKGDTMLLLLAAAHRDPTIVGAPDRFDPDRA
MLBr_02088|M.leprae_Br4923 MTIG-----------QTTLTEGDTMVLLLAAANRDPAVYSRPDEFDPDRP
MAB_0276|M.abscessus_ATCC_1997 MAIR-----------DFILPDGEWVLLMLAAAQRDPDVGPDLDVFNPDRS
TH_1650|M.thermoresistible__bu -------------------RRGRQ------GAGRDAHRTAR----NPGAR
Mb1912c|M.bovis_AF2122/97 VEVAG-----------VRIKRGEVVVIYLAAANRDPAVFPDPHRFDIERP
Rv1880c|M.tuberculosis_H37Rv VEVAG-----------VRIKRGEVVVIYLAAANRDPAVFPDPHRFDIERP
MUL_2979|M.ulcerans_Agy99 VEIAS-----------WPINRGEMVVAYLAGANRDPAVFPNPHRFDVERA
* .* **. :
MAV_0358|M.avium_104 SL-RHLGFGRGVHYCLGAPLARLEAGVALSAVTARFPRARLDGEPQYKTN
MMAR_5268|M.marinum_M AL-RHLGFGRGAHYCLGAPLARLEASVALSAIAARFPQARLDGEPQYKTN
Mflv_1229|M.gilvum_PYR-GCK SF-RHLGFGKGPHFCIGAPLARLEAAVALTALTSRFPGAQLAGEPSYKQN
Mvan_5579|M.vanbaalenii_PYR-1 VI-RHLGFGKGAHFCLGAPLARLEATLALTALTSRFPDAKLAAEPVYKPN
MSMEG_6312|M.smegmatis_MC2_155 QI-RHLGFGKGAHFCLGAPLARLEATVALPALAARFPEARLSGEPEYKRN
MLBr_02088|M.leprae_Br4923 SS-RHLAFAVGSHFCLGAALARLEATVTLSAISARFPQVQLAGELVYKPN
MAB_0276|M.abscessus_ATCC_1997 EI-KHLAFGHGPHFCLGAPLARLEARLALSSITARFPGARLMDEPTYKPN
TH_1650|M.thermoresistible__bu TV-RDVGR-------------RSDRRVRP----ARAAAAHRTGDGRVRTG
Mb1912c|M.bovis_AF2122/97 NAGRHLAFSTGRHFCLGAALARAEGEVGLRTFFDRFPDVRAAGAGSRRDT
Rv1880c|M.tuberculosis_H37Rv NAGRHLAFSTGRHFCLGAALARAEGEVGLRTFFDRFPDVRAAGAGSRRDT
MUL_2979|M.ulcerans_Agy99 NAGRHLAFSTGRHFCLGAALARAEGEVGLRTFFERFPDVRAAGAGSRRDT
:.:. * : : * . .: :
MAV_0358|M.avium_104 VTLRGLSRLVVAV---------
MMAR_5268|M.marinum_M VTLRGLESMTVALS--------
Mflv_1229|M.gilvum_PYR-GCK VTLRGMSELPITL---------
Mvan_5579|M.vanbaalenii_PYR-1 VTLRGMSELSITLA--------
MSMEG_6312|M.smegmatis_MC2_155 LTLRGMSTLSIAV---------
MLBr_02088|M.leprae_Br4923 VAMRGMSALPVQV---------
MAB_0276|M.abscessus_ATCC_1997 VTLRGLAELPVSIA--------
TH_1650|M.thermoresistible__bu PRAR------------------
Mb1912c|M.bovis_AF2122/97 RVLRGWSTLPVTLGPARSMVSP
Rv1880c|M.tuberculosis_H37Rv RVLRGWSTLPVTLGPARSMVSP
MUL_2979|M.ulcerans_Agy99 RVLRGWSTLPVTLGPARALAAS
*