For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VLTLPRLPRFLRSASLRTRVAIAAAAAAAAVVAAFTILTSLVLANNDEAQLDRRLDAIVDASMFPDQLSD PRRGVLQTGRSRSSGDVVFQRGFQLPPLPPGTDTVMVNGVEYRVRTIPVEQGGGVLMSIGIRADSILLSR ARIPFYVGVGVVTVLIAGVLGWLLAGPAIRPLRKLTEHTKRLDHETEQMPAVRGVREAEDLSEAMSAMLD RLAAAQRATTNSLQAAQDFAANAAHELRTPLTAMRADLDTLRIHNLPDDEREEVVADLSRAQRRVEGIIT ALGQLASGQLAQAEDREVIDVTDMLERVARENMRVRRDVQIDIDAADDLGTVLGWPGGLRLAVDNLVRNA ATHGEATRIVLAARRDGGAVTIVVDDNGRGLPADEHEAVLGRFSRGSNAAPGGSGLGLALVAQQAALHGG TITLSDGPLGGLRATFTISNEPAPQQDSDS
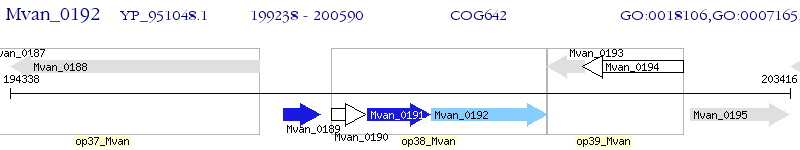
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0192 | - | - | 100% (450) | integral membrane sensor signal transduction histidine kinase |
| M. vanbaalenii PYR-1 | Mvan_5010 | - | 2e-77 | 38.91% (442) | integral membrane sensor signal transduction histidine kinase |
| M. vanbaalenii PYR-1 | Mvan_2118 | - | 2e-67 | 37.61% (436) | integral membrane sensor signal transduction histidine kinase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0926c | prrB | 8e-77 | 40.27% (437) | two component sensor histidine kinase PRRB |
| M. gilvum PYR-GCK | Mflv_0460 | - | 0.0 | 86.80% (447) | integral membrane sensor signal transduction histidine kinase |
| M. tuberculosis H37Rv | Rv0902c | prrB | 8e-77 | 40.27% (437) | two component sensor histidine kinase PRRB |
| M. leprae Br4923 | MLBr_02124 | - | 1e-76 | 39.33% (445) | sensor histidine kinase |
| M. abscessus ATCC 19977 | MAB_1927 | - | 1e-127 | 57.41% (432) | putative sensor-type histidine kinase PrrB |
| M. marinum M | MMAR_4634 | prrB | 4e-76 | 40.50% (437) | two-component sensor histidine kinase PrrB |
| M. avium 104 | MAV_1021 | - | 5e-81 | 39.86% (434) | sensor-type histidine kinase PrrB |
| M. smegmatis MC2 155 | MSMEG_0246 | - | 0.0 | 78.10% (443) | sensor-type histidine kinase PrrB |
| M. thermoresistible (build 8) | TH_0454 | - | 2e-80 | 40.31% (449) | sensor-type histidine kinase PrrB |
| M. ulcerans Agy99 | MUL_0249 | prrB | 7e-76 | 40.27% (437) | two component sensor histidine kinase PrrB |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_4634|M.marinum_M MTVMNILSRIFARTPSLRTRVVVATAIGAAIPVLIVGTVVWVGITNDRKE
MUL_0249|M.ulcerans_Agy99 ---MNILSRIFARTPSLRTRVVVATAIGAAIPVLIVGTVVWVGITNDRKE
Mb0926c|M.bovis_AF2122/97 ---MNILSRIFARTPSLRTRVVVATAIGAAIPVLIVGTVVWVGITNDRKE
Rv0902c|M.tuberculosis_H37Rv ---MNILSRIFARTPSLRTRVVVATAIGAAIPVLIVGTVVWVGITNDRKE
MLBr_02124|M.leprae_Br4923 ---MNILSRIFARTPSLRTRVVVATAIGAAIPVLIVGTVVWVGITNDRKE
MAV_1021|M.avium_104 ---MKLLSRIVTRTPSLRARVAVATAIGTAIVVVIAGAVVWFGITSAWRE
TH_0454|M.thermoresistible__bu ---MNLVTRLFRRTPSLRTRVAFATAIAAAIVVGIVGVVVWVGITNDRKE
Mvan_0192|M.vanbaalenii_PYR-1 -MLTLPRLPRFLRSASLRTRVAIAAAAAAAAVVAAFTILTSLVLANNDEA
Mflv_0460|M.gilvum_PYR-GCK ----MPRLPRFLSSASLRTRVAIASAAAAAAVVAAFTILTSLVLANNDAA
MSMEG_0246|M.smegmatis_MC2_155 --MRRLRLPAFIRSASLRTRVAVAAAAAASAVVAAFTILTSVVLASNDAA
MAB_1927|M.abscessus_ATCC_1997 ----------MKRSLSLRTRVAVAAFLAAGLVVALIGIVAGIALVRNDTR
. : ***:**..*: .:. * :. . :.
MMAR_4634|M.marinum_M RLDRRLDEAAGFAIPFVPRGLDEIPRSPNDQDALITVRRGDLVKSNSDIT
MUL_0249|M.ulcerans_Agy99 RLDRRLDEAAGFAIPFVPRGLDEIPRSPNDQDALITVRRGDLVKSNSDIT
Mb0926c|M.bovis_AF2122/97 RLDRRLDEAAGFAIPFVPRGLDEIPRSPNDQDALITVRRGNVIKSNSDIT
Rv0902c|M.tuberculosis_H37Rv RLDRRLDEAAGFAIPFVPRGLDEIPRSPNDQDALITVRRGNVIKSNSDIT
MLBr_02124|M.leprae_Br4923 RLDRKLDEAAGFAIPFVPRGLDEIPRSPNDQDAIITVRRGNLVKSNFDIT
MAV_1021|M.avium_104 RLDRRLDETAGFAIPFLPRGLNEIPRSPKDQDTVITVRRDGQVKSNSDVT
TH_0454|M.thermoresistible__bu RLDRRLDEAAGFALPFVPRGLEQIPPSPNDQDVVITVRRGDEVRSNSEVV
Mvan_0192|M.vanbaalenii_PYR-1 QLDRRLDAIVDASM-----FPDQLSDPRRGVLQTGRSRSSGDVVFQRGFQ
Mflv_0460|M.gilvum_PYR-GCK QLDRRLDAIVDASV-----FPDQLTDPRRGVLQTGRSRSSGQVVFQRGFQ
MSMEG_0246|M.smegmatis_MC2_155 QLDRRLDAIVDASI-----NPEHLQDPQRGVLTTGRTRTGGQVVFQRGFQ
MAB_1927|M.abscessus_ATCC_1997 QLDRRLDMLTDAVQ-----ATTRHGETT-FVLTVRDTKSGGLVIFR-GVE
:***:** .. . . : .. : . .
MMAR_4634|M.marinum_M LPKLQKDYADTYIRGVRYRVRTVEIPGPEPTSVAVGATYDATIAETNNLH
MUL_0249|M.ulcerans_Agy99 LPKLQEDYADTYIRGVRYRVRTVEIPGPEPTSVAVGATYDATIAETNNLH
Mb0926c|M.bovis_AF2122/97 LPKLQDDYADTYVRGVRYRVRTVEIPGPEPTSVAVGATYDATVAETNNLH
Rv0902c|M.tuberculosis_H37Rv LPKLQDDYADTYVRGVRYRVRTVEIPGPEPTSVAVGATYDATVAETNNLH
MLBr_02124|M.leprae_Br4923 LPKLTNDYADTYLRGVRYRVRTVEIPAPEPTSIAVGATYDATVAETNNLH
MAV_1021|M.avium_104 LPPMEEGYADTYINGVRYRVRTVEVRTPQPATVEVGATYDDTIAQTNNLH
TH_0454|M.thermoresistible__bu LPELDADYADTYIDGVRYRVRTVQTPYPIRTTLAVGATYDDTIADTANLH
Mvan_0192|M.vanbaalenii_PYR-1 LPPLPPGTDTVMVNGVEYRVRTIPVEQGGGVLMSIGIRADSILLSRARIP
Mflv_0460|M.gilvum_PYR-GCK LPPLPPGTETVIVNGVEYRVRTIPVDEGGGVLMSIGIRADSILLSAARVP
MSMEG_0246|M.smegmatis_MC2_155 LPALPPGTATVEVNGVDYRVRTVSVDQQGGIMMSIGIRADSILLSANRIP
MAB_1927|M.abscessus_ATCC_1997 LPRLPLGTRNVTVGDVNYRVRTIENPEEQ-TVISLGTRSDTVLVNHRGIP
** : . . : .* *****: : :* * : . :
MMAR_4634|M.marinum_M RRVLLICTFAIGAATVFAWLLAVFAVRPFKQLAEQTRSIDAGDEAPRVEV
MUL_0249|M.ulcerans_Agy99 RRVLLICTFAIGAATVFAWLLAVFAVRPFKQLAEQTRSIDAGDEAPRVEV
Mb0926c|M.bovis_AF2122/97 RRVLLICTFAIGAAAVFAWLLAAFAVRPFKQLAEQTRSIDAGDEAPRVEV
Rv0902c|M.tuberculosis_H37Rv RRVLLICTFAIGAAAVFAWLLAAFAVRPFKQLAEQTRSIDAGDEAPRVEV
MLBr_02124|M.leprae_Br4923 RRVLLICGFAIAAAAVFAWLLAAFAVRPFKQLAQQTRSVDAGGEAPRVEV
MAV_1021|M.avium_104 RRVILICALSIGAAAVFAWLLAAFAVRPFKRLAQQARSIDA-DERPQVAV
TH_0454|M.thermoresistible__bu RRVILICTLAIGAATIAGWLLAAFAVRPFKQLAAQARSIDAGDEVPDVEI
Mvan_0192|M.vanbaalenii_PYR-1 -FYVGVGVVTVLIAGVLGWLLAGPAIRPLRKLTEHTKRLD-HETEQMPAV
Mflv_0460|M.gilvum_PYR-GCK -FYIGVGVVTVLLAGVLGWLLAGPAIRPLRKLTEHTKQLG-DGAEQIPAV
MSMEG_0246|M.smegmatis_MC2_155 -LYAAVGVVTVLVAAGLGWLLAGPAIRPLRKLTEHTSKLGSGGPDTLPEV
MAB_1927|M.abscessus_ATCC_1997 -VFIALGVLSMLLAGGFAWLFAGRAVRPLHQLTDQTRQLD-TDGLQVAAV
: .:: * .**:* *:**:::*: :: :. :
MMAR_4634|M.marinum_M HGASEAIEIAEAMRGMLQRIWNEQIRTKEALASARDFAAVSSHELRTPLT
MUL_0249|M.ulcerans_Agy99 HGASEAIEIAEGMRGMLQRIWNEQIRTKEALASARDFAAVSSHELRTPLT
Mb0926c|M.bovis_AF2122/97 HGASEAIEIAEAMRGMLQRIWNEQNRTKEALASARDFAAVSSHELRTPLT
Rv0902c|M.tuberculosis_H37Rv HGASEAIEIAEAMRGMLQRIWNEQNRTKEALASARDFAAVSSHELRTPLT
MLBr_02124|M.leprae_Br4923 HGATEAVEIAEAMRGMLQRIWNEQNRTKEALASARDFAAVSSHELRTPLT
MAV_1021|M.avium_104 RGASEAVEIAEAMRGMLQRIWKEQDRTKEALASARDFSAVTAHELRSPLT
TH_0454|M.thermoresistible__bu RGATEAVEIADAVKGMLQRIWKEQDRTKAALASARDFASAAAHELRTPLT
Mvan_0192|M.vanbaalenii_PYR-1 RGVREAEDLSEAMSAMLDRLAAAQRATTNSLQAAQDFAANAAHELRTPLT
Mflv_0460|M.gilvum_PYR-GCK RGVREAEDLSEAMSAMLDRLAAAQRVTTNSLQAAQDFAANAAHELRTPLT
MSMEG_0246|M.smegmatis_MC2_155 RGVREAEDLSEAMSGMLTRLAAAQRATTNSLQAAQDFAANAAHELRTPLT
MAB_1927|M.abscessus_ATCC_1997 RGARESEELAEAMAAMLSRVSAAQSETTKSLLAARDFAASAAHELRTPLT
:*. *: ::::.: .** *: * *. :* :*:**:: ::****:***
MMAR_4634|M.marinum_M AMRTNLEVLSTLDLPDDQRKEVLNDVIRTQSRIEATLGALERLAQGELST
MUL_0249|M.ulcerans_Agy99 AMRTNLEVLSTLDLPDDQRKEVLNDVIRTQSRIEATLGALERLAQGELST
Mb0926c|M.bovis_AF2122/97 AMRTNLEVLSTLDLPDDQRKEVLNDVIRTQSRIEATLSALERLAQGELST
Rv0902c|M.tuberculosis_H37Rv AMRTNLEVLSTLDLPDDQRKEVLNDVIRTQSRIEATLSALERLAQGELST
MLBr_02124|M.leprae_Br4923 AMRTNLEVLATLDLADDQRKEVLGDVIRTQSRIEATLSALERLAQGELST
MAV_1021|M.avium_104 AMRTNLEVLSTLDMPDEQRKEVLGDVIRTQSRIEATLTALERLAQGELST
TH_0454|M.thermoresistible__bu AMRTNLEVLSTLDMPEEQRKEVLADVIRTQSRIEATLSALERLAQGELST
Mvan_0192|M.vanbaalenii_PYR-1 AMRADLDTLRIHNLPDDEREEVVADLSRAQRRVEGIITALGQLASGQLAQ
Mflv_0460|M.gilvum_PYR-GCK AMRADLDTLRIHNLPDEERDEVVADLARAQRRVEGIITALGQLASGQLAQ
MSMEG_0246|M.smegmatis_MC2_155 AMRADLDTLRIHDLPEDERDEVVADLSRAQRRVEAIITALGQLASGQLAQ
MAB_1927|M.abscessus_ATCC_1997 AMRADLDTLRAHDLSGDERVEVIGDLQRAQRRVEATITALGQLASGDLAR
***::*:.* ::. ::* **: *: *:* *:*. : ** :**.*:*:
MMAR_4634|M.marinum_M SEDHVPVDITELLDRAAHDAMRIYPDLDVSLVPSPTC-IIVGLPAGLRLA
MUL_0249|M.ulcerans_Agy99 SEDHVPVDITELLDRAAHDAMRIYPDLDVSLVPSPTC-IIVGLPAGLRLA
Mb0926c|M.bovis_AF2122/97 SDDHVPVDITDLLDRAAHDAARIYPDLDVSLVPSPTC-IIVGLPAGLRLA
Rv0902c|M.tuberculosis_H37Rv SDDHVPVDITDLLDRAAHDAARIYPDLDVSLVPSPTC-IIVGLPAGLRLA
MLBr_02124|M.leprae_Br4923 SDDHVPVDITELLDRAAHDATRSYPELKVSLVPSPTC-IIVGLPAGLRLA
MAV_1021|M.avium_104 SEDHVPVDITELLDRAAHDAMRIYPGLDVSLVPSPTC-IIVGLPAGLRLA
TH_0454|M.thermoresistible__bu SDDHVPVDITELLDRAAHEAMRIYPDLEVSLVPAPTV-IIVGLPAGLRLA
Mvan_0192|M.vanbaalenii_PYR-1 AEDREVIDVTDMLERVARENMRVRRDVQIDIDAADDLGTVLGWPGGLRLA
Mflv_0460|M.gilvum_PYR-GCK VEDREQIDVADLLDRVARENMRVSRDVQIDIDAVDDLGSVQGWPSGLRLA
MSMEG_0246|M.smegmatis_MC2_155 AEDRELIDITDMLDRVARENTRADTGVRIEVHADDDVGLVLGWPGGLRIA
MAB_1927|M.abscessus_ATCC_1997 TEDRELVDIADLLDRVAGDALMHRADAPEITVHADDCGVVLGWPDGLRLA
:*: :*::::*:*.* : : * * ***:*
MMAR_4634|M.marinum_M VDNAIANAVKHGGSTRVQLSAVSSRAGVEIAVDDNGSGVPEAERQMVFER
MUL_0249|M.ulcerans_Agy99 VDNAIANAVKHGGSTRVQLSAVSSRAGVEIAVDDNGSGVPEAERQMVFER
Mb0926c|M.bovis_AF2122/97 VDNAIANAVKHGGATLVQLSAVSSRAGVEIAIDDNGSGVPEGERQVVFER
Rv0902c|M.tuberculosis_H37Rv VDNAIANAVKHGGATLVQLSAVSSRAGVEIAIDDNGSGVPEGERQVVFER
MLBr_02124|M.leprae_Br4923 VDNAVANAVKHGGATRVQLSAVSSRAGVEIAVDDNGSGVPEDERQVVFER
MAV_1021|M.avium_104 VDNAIANAVKHGGATRVQLSAVSSRAGVEIAVDDNGSGVPEDERQVVFDR
TH_0454|M.thermoresistible__bu VDNAIANAVKHGGATRVQLSAVSSREGVEIAIDDNGTGVPEEERERVFDR
Mvan_0192|M.vanbaalenii_PYR-1 VDNLVRNAATHGEATRIVLAARRDGGAVTIVVDDNGRGLPADEHEAVLGR
Mflv_0460|M.gilvum_PYR-GCK VDNLVRNAVTHGQASQIVLSARREDSAMTIVVDDNGRGLPADEHQTVLGR
MSMEG_0246|M.smegmatis_MC2_155 VDNLVRNAITHGEAGRVVVSAHRTGQMLTIVVDDNGRGLPVEEHETVLGR
MAB_1927|M.abscessus_ATCC_1997 VDNLVRNAINHGEARHIVLSANRIDNRLVIEVDDDGPGLPEADRERVLGR
*** : ** .** : : ::* : * :**:* *:* ::: *: *
MMAR_4634|M.marinum_M FSRGSTASHSGSGLGLALVAQQAHLHGGTASLQNSPLGGARLLLKLPGPS
MUL_0249|M.ulcerans_Agy99 FSRGSTASHSGSGLGLALVAQQAHLHGGTASLQNSPLGGARLLLKLPGPS
Mb0926c|M.bovis_AF2122/97 FSRGSTASHSGSGLGLALVAQQAQLHGGTASLENSPLGGARLVLRLPGPS
Rv0902c|M.tuberculosis_H37Rv FSRGSTASHSGSGLGLALVAQQAQLHGGTASLENSPLGGARLVLRLPGPS
MLBr_02124|M.leprae_Br4923 FSRGSTASHSGSGLGLALVAQQAQLHGGTASLETSPLGGARLLLRISAPS
MAV_1021|M.avium_104 FSRGSTASQSGSGLGLALVAQQAQLHGGTAALENSPLGGARLVLRLPGPS
TH_0454|M.thermoresistible__bu FTRGSTASHSGSGLGLALVAQQAEIHGGTASLEASPLGGARLVLRLPGPR
Mvan_0192|M.vanbaalenii_PYR-1 FSRGSNAAPGGSGLGLALVAQQAALHGGTITLSDGPLGGLRATFTISNEP
Mflv_0460|M.gilvum_PYR-GCK FSRGSTAAPGGSGLGLALVAQQAALHGGTIELTDSPLGGLRATLTISTSL
MSMEG_0246|M.smegmatis_MC2_155 FRRGSTAVAGGSGLGLALVAQQAELHGGQIKLSDGPLGGLRATLTVSIAP
MAB_1927|M.abscessus_ATCC_1997 FERGPGAKPGGLGLGLALVAQQAALHGGDVVLSSSPLGGLRATLTIAAE-
* **. * .* *********** :*** * .**** * : :.
MMAR_4634|M.marinum_M --------
MUL_0249|M.ulcerans_Agy99 --------
Mb0926c|M.bovis_AF2122/97 --------
Rv0902c|M.tuberculosis_H37Rv --------
MLBr_02124|M.leprae_Br4923 --------
MAV_1021|M.avium_104 --------
TH_0454|M.thermoresistible__bu --------
Mvan_0192|M.vanbaalenii_PYR-1 APQQDSDS
Mflv_0460|M.gilvum_PYR-GCK VPLGDPES
MSMEG_0246|M.smegmatis_MC2_155 TGPTDQLA
MAB_1927|M.abscessus_ATCC_1997 --------