For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAAETPALAPPPKVTEVETHGVEPIPDAERSARPLDLFRLTFGGANTIATVVLGSFPIIFGLSFRDALLA TLLGVVAGAAILAPMALFGPRNGTNNAVSSSAHLGVHGRVVGSFLSLLTAVAFFSISVWTSGDVLVGGAT RAIGGAGSAAPNDWAVGVAYGVFALLVLTVCIYGFRFMLLVNKVAVVAATVLFLLGIFAFGGTFDPGYAG IFGPDADAATNALYWPSFVGAALIVMSNPVSFGAFLGDWARYIPRDTPPWKPMAAAFCAQLATLVPFLFG LVTAAVVATNAPQFIDEGNYVGGLLEVSPGWYFLPVCLIALIGGMSTGTTALYGTGLDFSSVFPRFSRVQ ATVFIGVLAIVFIFIGRFAFNVVQSISTFAVLIVTCTAPWMVVMTTGWVVRRGWYDSDSLQVFNRRQRGG RYWFARGWNFRGLGAWLVSAALSLCFVNLPGQFVGPLGDLADGIDLSIPVGLGVAALLYPLLLVAFPEPA DAFGPEGPRFVRAGAPAGIPITSEDEAEKPVVSEGVTA
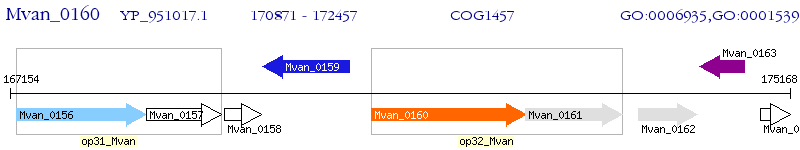
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0160 | - | - | 100% (528) | permease for cytosine/purines, uracil, thiamine, allantoin |
| M. vanbaalenii PYR-1 | Mvan_3383 | - | 2e-21 | 25.68% (475) | permease for cytosine/purines, uracil, thiamine, allantoin |
| M. vanbaalenii PYR-1 | Mvan_0862 | - | 1e-15 | 23.26% (374) | permease for cytosine/purines, uracil, thiamine, allantoin |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_0055 | - | 9e-16 | 21.95% (369) | permease for cytosine/purines, uracil, thiamine, allantoin |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2025 | - | 1e-78 | 32.99% (479) | purine/cytosine permease |
| M. marinum M | MMAR_2052 | - | 2e-07 | 23.47% (392) | amino acid transporter |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_4458 | - | 0.0 | 81.48% (513) | transmembrane transport protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_0160|M.vanbaalenii_PYR-1 -----------MAAETPALAPPPKVTEVETHGVEPIPDAERSARPLDLFR
MSMEG_4458|M.smegmatis_MC2_155 -----------MATTEPVPVTP-RVTEVEQHGVEPIPDAERTARPLDLFR
MAB_2025|M.abscessus_ATCC_1997 MDEIQACCSATSLIASRESVGRDRVGAIERAGIDYLPEAARGSSPKNLFA
Mflv_0055|M.gilvum_PYR-GCK -------------------------MHDLEAELTVIPESAKTTDLRGQFW
MMAR_2052|M.marinum_M ----------------------MSEGPQRRAEAAPATLAASALRPRNVLA
. : . :
Mvan_0160|M.vanbaalenii_PYR-1 LTFGGANTIATVVLGSFPIIFGLSFRDALLATLLGVVAGAAILAPMALFG
MSMEG_4458|M.smegmatis_MC2_155 LVFGGANTIATVVLGTFPIIFGLSFRDALLATLAGLVLGALILAPMSLFG
MAB_2025|M.abscessus_ATCC_1997 VFVGGNLAFSTIIFGWLPIVMGLTFWQAVSSSVTGLLIGLMLTAPMSLFG
Mflv_0055|M.gilvum_PYR-GCK IWAGANIAPINWILGALGVTMGLGLFETIAVLVVGNAIGMSLFGTFVVIG
MMAR_2052|M.marinum_M IAVSAISPTTSVFL-----IYGDGLQIAGTGVVWAFVIGSAIAVCMALCY
: .. . . .: * : : : . * : : :
Mvan_0160|M.vanbaalenii_PYR-1 PRNGTNNAVSSSAHLGVHGRVVGSFLSLLTAVAFFSISVWTSGDVLVGGA
MSMEG_4458|M.smegmatis_MC2_155 PRNGTNNAVSSSAHLGVHGRVVGSFLSLLTAVAFFSISVWTSGDVLVGGA
MAB_2025|M.abscessus_ATCC_1997 PRTGTNNPVSSGAHFGVRGRLIGSSLTLMFALVYAAIAVWTSGEALIAGA
Mflv_0055|M.gilvum_PYR-GCK QKTGVTGMVLSRMVFGRRGAYVPAAVQAALCIGWCAVNTWIVLDLVMALL
MMAR_2052|M.marinum_M AEVGSVFPSAGGAYTIVR-RAIGPVAGGVTSVLFLLLGVVSTAAILVAAA
. * . : : . .: : : . ::.
Mvan_0160|M.vanbaalenii_PYR-1 TRAIGGAGSAAPNDWAVGVAYGVFALLVLTVCIYGFRFMLLVNKVAVV--
MSMEG_4458|M.smegmatis_MC2_155 NRAID----VPQNDIAVGVAYGIFAVLVLVVCIYGFRFMLLVNKIAVI--
MAB_2025|M.abscessus_ATCC_1997 SRLFG----APQNHVAFLLGYGVIALEVILIALYGHATIVWAQKIIIP--
Mflv_0055|M.gilvum_PYR-GCK G-RIGLVDPELSNHAARIVVAGIIMTTQVVIAWFGYRLIAAFERWTVPPT
MMAR_2052|M.marinum_M TYLSS---LIGAAVPVRWLALGMMALITALSIGKIAPASWVVAAMLALEL
. . : *::
Mvan_0160|M.vanbaalenii_PYR-1 AATVLFLLGIFAFGGTFDPGYAGIFGPDADAATNALYWPSFVGAALIVMS
MSMEG_4458|M.smegmatis_MC2_155 AATLLFLAGVFAFGGVFDAGYAG-----SVALGDPLFWPSFVGAALIVMS
MAB_2025|M.abscessus_ATCC_1997 LAVVVLALGVLAYAPNFHPMGSA-----QHGYALQEFWPTWLFAVTVSVG
Mflv_0055|M.gilvum_PYR-GCK VAVLVAMTAVAWFGLDVDWGYTG----DAALTGAEHFTAITAVMTAIGIG
MMAR_2052|M.marinum_M AVIIVFTVVAFAHASAVTKPFTQ-----PVTAGPGGALVGVGIAGIFAAV
. :: .. . : .
Mvan_0160|M.vanbaalenii_PYR-1 NPVSFGAFLGDWARYIPRDTPPWKPMAAAFCAQLATLVPFLFGLVTAAVV
MSMEG_4458|M.smegmatis_MC2_155 NPVSFGAFLGDWARYIPRDTPSWKPMLAAFSAQIATLVPFLFGLVTATVI
MAB_2025|M.abscessus_ATCC_1997 GPLSYAPSVGDYTRRISRSRYSDRQIAVAVSAG-----IFLGLFSTATFG
Mflv_0055|M.gilvum_PYR-GCK WGITWLGYAGDYSRFVSSSVPARKLFAASALGQ------FIPVVWLGALG
MMAR_2052|M.marinum_M GPALFAFNGYDWPLYFSEETAGSRRAIPHAVVLS-----AVLAVVVECVA
: *:. .. . : : . .
Mvan_0160|M.vanbaalenii_PYR-1 ATNAPQFID-EGNYVGGLLEVSPGWYFLPVCLIALIGG---MSTGTTALY
MSMEG_4458|M.smegmatis_MC2_155 ATTAPDYIA-NGDYVGGLLSVSPGWYFVPVCLIALIGG---MSTGTTALY
MAB_2025|M.abscessus_ATCC_1997 AFTASTFSAPTASYVADMVATAPGWYVVPLLILAVLGG---CGQGVISIY
Mflv_0055|M.gilvum_PYR-GCK ATLATLSAS---SDPGEIIVDAYGVLAIPVLLLVVHGP---LATNILNIY
MMAR_2052|M.marinum_M VIAATLAIR----DIPAAITDGSAVEFLARQMMGPAGATVLVAGVVVAMF
. *. : . . :. :: * . ::
Mvan_0160|M.vanbaalenii_PYR-1 GTGLDFSSVFPRFSRVQATVFIGVLAIVFIFIGRFAFNVVQSISTFAVLI
MSMEG_4458|M.smegmatis_MC2_155 GTGLDFSSVFPKFSRVQATIFIGSIAIVFIFIGRFAFNVVQSISTFAVLI
MAB_2025|M.abscessus_ATCC_1997 SSGLDLESIVPRLNRTQTTLIASGIAVVLMYLGVVVTDAIDSVTGMTVVL
Mflv_0055|M.gilvum_PYR-GCK TCAVSTQALDIHVNRRVLNIVIGVVAMAWVVYFVLNGDFAATIDAWLVGI
MMAR_2052|M.marinum_M DTGLAANLGYARVYYAAARDGMLPRRLQPLFG-RLSTRSRVPVHGFLFLF
.: . :. : : . .: . :
Mvan_0160|M.vanbaalenii_PYR-1 VTCTAPWMVVMTTGWVVR-RGWYDSDSLQVFNRRQRGGRYWFARGWNFRG
MSMEG_4458|M.smegmatis_MC2_155 VTCTAPWMVVMMIGWFTR-RGWYDSDALQVFNRRQRGGRYWFDHGWNWRG
MAB_2025|M.abscessus_ATCC_1997 NSFAGSWVAINICGFFKANRGQYDPKALQRFGQDGRGGLYWFSHGFNVRA
Mflv_0055|M.gilvum_PYR-GCK VGWLAPWAAVMLVHWFGVARRQVDPATLLTPPNES------TLPAVRPSA
MMAR_2052|M.marinum_M LG-NGLLCVFMSLPKLITFTGVVIAIVYLLVALSAIVCRLRDRDLVRPFR
. .. . . .
Mvan_0160|M.vanbaalenii_PYR-1 LGAWLVSAALS-LCFVNLPGQFVGPLGDLADGIDLSIPVGLGVAALLYPL
MSMEG_4458|M.smegmatis_MC2_155 LTAWLISAALS-ICFVNLPDQFVGPLGNLADGIDLSIPVGLGLAAVLYPA
MAB_2025|M.abscessus_ATCC_1997 VVPFIAGSGLG-LLTVQN-DLYQAPFANIAGGIDVSLLLSVVVGGGLYWL
Mflv_0055|M.gilvum_PYR-GCK LISLAVGIVFTWLFMYGMVPMMQGPIAVALGGVDLSWLAGGLAAGSLYAA
MMAR_2052|M.marinum_M MPLWPLPPLLAVVGVVVTLATQEVKDLAIGAGLAAAAALGYRIARRRLPA
: : : *: : . .
Mvan_0160|M.vanbaalenii_PYR-1 LLVAFPEPADAFGPEGPRFVRAGAPAGIPITSEDEAEKPVVSEGVTA
MSMEG_4458|M.smegmatis_MC2_155 LLWLSPEPRDAFGPDGPRGVPSGAPANTPIESQD-TVISTHTEEVPA
MAB_2025|M.abscessus_ATCC_1997 AQVSWP---------APRVVSSGSSA---------------------
Mflv_0055|M.gilvum_PYR-GCK LEIPKARRA--------------------------------------
MMAR_2052|M.marinum_M RLGGDP-------------VTGRSTP---------------------
.