For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NAPSLVNHLVAPFGMQGFSYLPSVGILSTYVPTACGLATFSAALAGGLSAKGISVNVVRIADGTPSSSAD VVGELVNGSASSVAAASESLNSNDIAVIQHEYGIYGGADGDEVVDIIRGLRVPAIVVVHTVLKTPTPHQR SVLEEVLALAGCAVVMSEAARSLLCAGYAVDPQKVIIIPHGAAPAEDVEWKPAVRPTILTWGLLGPGKGV ERVIDVMASLQDLPDPPRYLIAGRTHPKVLAAQGEAYRDGLAARADRLGVGDSVIFDATYRSPQALAEML RDAAVVVLPYDSKDQVTSGVLIDSIANGRPVIATAFPHAVEVLGNGAGTVVDHDDPEALTDALRRLITDP DTAGSMAAVARTLAPSVAWPVVADAYAALAQRLVAEKLATA*
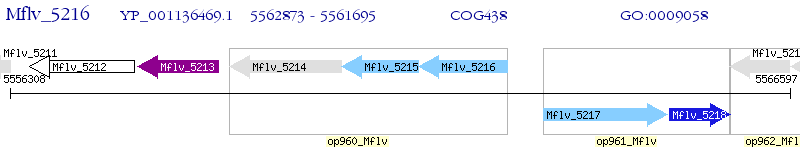
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_5216 | - | - | 100% (392) | glycosyl transferase, group 1 |
| M. gilvum PYR-GCK | Mflv_5224 | - | e-120 | 57.52% (379) | glycosyl transferase, group 1 |
| M. gilvum PYR-GCK | Mflv_0426 | - | 4e-08 | 24.68% (235) | glycosyl transferase, group 1 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0496 | - | 9e-08 | 27.53% (287) | mannosyltransferase |
| M. tuberculosis H37Rv | Rv0486 | - | 9e-08 | 27.53% (287) | mannosyltransferase |
| M. leprae Br4923 | MLBr_00886 | - | 3e-09 | 27.41% (259) | putative glycosyl transferase |
| M. abscessus ATCC 19977 | MAB_1976 | - | 3e-08 | 27.75% (173) | hypothetical protein MAB_1976 |
| M. marinum M | MMAR_4514 | - | 3e-07 | 24.46% (278) | glycosyl transferase |
| M. avium 104 | MAV_4664 | - | 2e-08 | 26.95% (282) | hypothetical protein MAV_4664 |
| M. smegmatis MC2 155 | MSMEG_1207 | - | 1e-128 | 64.50% (369) | glycosyltransferase, group I |
| M. thermoresistible (build 8) | TH_1111 | - | 6e-07 | 27.32% (194) | CONSERVED HYPOTHETICAL PROTEIN |
| M. ulcerans Agy99 | MUL_1124 | - | 9e-08 | 22.03% (236) | glycosyltransferase |
| M. vanbaalenii PYR-1 | Mvan_1052 | - | 1e-163 | 78.44% (371) | glycosyl transferase, group 1 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0496|M.bovis_AF2122/97 MAGVRHDDGSGLIAQRRPVRGEGATRSRGPSGPSNRNVSAADDPRRVALL
Rv0486|M.tuberculosis_H37Rv MAGVRHDDGSGLIAQRRPVRGEGATRSRGPSGPSNRNVSAADDPRRVALL
MAV_4664|M.avium_104 ---MRHDD-----------------------------VFRLNEPRRVAVL
MAB_1976|M.abscessus_ATCC_1997 -------------------------------------MTFHPGGLRQRIL
TH_1111|M.thermoresistible__bu ----------------------------------------VTG--VSRVL
MLBr_00886|M.leprae_Br4923 ---------------------------------------------MSRVL
Mflv_5216|M.gilvum_PYR-GCK -------------------------MNAPSLVNHLVAPFGMQGFSYLPSV
Mvan_1052|M.vanbaalenii_PYR-1 ----------------------------------------------MPSV
MSMEG_1207|M.smegmatis_MC2_155 -------------------------MTTASAWPLTPGVATRQGCSDGLNV
MUL_1124|M.ulcerans_Agy99 ----------------------------------------MSDLRSVLLL
MMAR_4514|M.marinum_M --------------------------------MNSVGVPTPQDRLRVLVV
:
Mb0496|M.bovis_AF2122/97 AVHTSPLAQPGTGDAGGMNVYMLQSALHLAR-RGIEVEIFTRATASADPP
Rv0486|M.tuberculosis_H37Rv AVHTSPLAQPGTGDAGGMNVYMLQSALHLAR-RGIEVEIFTRATASADPP
MAV_4664|M.avium_104 AVHTSPLAQPGTGDAGGMNVYVLQTALHLAR-RGIEVEIFTRATASADPP
MAB_1976|M.abscessus_ATCC_1997 LVTNDFPPRP-----GGIQSYLQEMVSRLAG--SHEVTVYAPRWKGCER-
TH_1111|M.thermoresistible__bu LVTNDFPPRP-----GGIQSYLHELVLRLVRTQSHVVTVYAPRWRGSDD-
MLBr_00886|M.leprae_Br4923 LVTNDFPPRR-----GGIQSYLGEFVDRLINIGSHDVMVYAPQWTGADT-
Mflv_5216|M.gilvum_PYR-GCK GILSTYVPTA-----CGLATFSAALAGGLSA---KGISVNVVRIADGTP-
Mvan_1052|M.vanbaalenii_PYR-1 GILSTYAPTV-----CGLATFSAALAGGLSA---RGIDVNVVRIADGTP-
MSMEG_1207|M.smegmatis_MC2_155 GILSTYAPTA-----CGLATFSAALAGGLSG---TGANVNVVRIADGAP-
MUL_1124|M.ulcerans_Agy99 CWRDTGHPQG-----GGSETYLQRIGAQLAA-SGIEVTLRTARYPGAPR-
MMAR_4514|M.marinum_M GPAPAAATSR-----GGMATVIALMAAHPDP----HIHITAVPT------
. * : .
Mb0496|M.bovis_AF2122/97 VVRVAPGVLVRNVVAGPFEGLDKYDLPTQLCAFAAGVLRAEAVHEPGY-Y
Rv0486|M.tuberculosis_H37Rv VVRVAPGVLVRNVVAGPFEGLDKYDLPTQLCAFAAGVLRAEAVHEPGY-Y
MAV_4664|M.avium_104 VQRVVPGVVVRNVVAGPFEGLDKYDLPTQLCAFAAGVLRAEASHEPGY-Y
MAB_1976|M.abscessus_ATCC_1997 ------------------YDAAAE---YQVVRHPTSLMLPGPGVRRRM-V
TH_1111|M.thermoresistible__bu ------------------FDAEAARTGYRVVRHPGTLMLPEPSVARRM-R
MLBr_00886|M.leprae_Br4923 ------------------FDNAAHDGGYQVVRHPGTLMLPGPAVDARM-R
Mflv_5216|M.gilvum_PYR-GCK -------------------SSSADVVGELVNGSASSVAAASESLNSND-I
Mvan_1052|M.vanbaalenii_PYR-1 -------------------SVDGKVVGELVNGSSPSVAAAAERLNGCD-V
MSMEG_1207|M.smegmatis_MC2_155 -------------------SGNARVVGELVNGSAASTAACVELLNQNH-V
MUL_1124|M.ulcerans_Agy99 -----------------WEVVDGVRVNRAGGRYSVYVWALLAMVAARMGL
MMAR_4514|M.marinum_M -------------------FIDGPLWPRLMLGVRGMLRATWLVLRKRADV
Mb0496|M.bovis_AF2122/97 DIVHSHYWLSGQVGWLARDRWAVPLVHTAHTLAAVKNAALADGDGPEPPL
Rv0486|M.tuberculosis_H37Rv DIVHSHYWLSGQVGWLARDRWAVPLVHTAHTLAAVKNAALADGDGPEPPL
MAV_4664|M.avium_104 DIVHSHYWLSGQVGWLARDRWAVPLVHTAHTLAAVKNAALAQGDSPEPPL
MAB_1976|M.abscessus_ATCC_1997 DLIRAQ---HADVVWFGAAAPLALLSSAAKAAGAAVTAASTHGHEVGWSM
TH_1111|M.thermoresistible__bu RLIRDE---GIDTVWFGAAAPLALLAGTARAAGARRVLASTHGHEVGWSM
MLBr_00886|M.leprae_Br4923 RLIVDH---GIDTVWFGSAAPLALLAPCARKAGATRVLASTHGHEVGWSM
Mflv_5216|M.gilvum_PYR-GCK AVIQHEY------GIYGGADGDEVVDIIRGLRVPAIVVVHTVLKTPTPHQ
Mvan_1052|M.vanbaalenii_PYR-1 AVIQHEY------GIYGGADGEEVVEIIRGLRVPSIVVVHTVLESPTPHQ
MSMEG_1207|M.smegmatis_MC2_155 AIVQHEY------GIYGGADGDEVVGIIGALDVPVIVVLHTVPSAPTAHQ
MUL_1124|M.ulcerans_Agy99 GPLQRVR----PAAVIDTQNGLPFLARLVYGRRVVVLVHHCHREQWPVAG
MMAR_4514|M.marinum_M LHVHLAH------GGSVIRKALPLLAARLTGTPAIVHAHSYDFGGWFDQL
: :
Mb0496|M.bovis_AF2122/97 RTVG--EQQVVDEADRLIVNTDDEARQVISLHGADPARIDVVHPGVDLDV
Rv0486|M.tuberculosis_H37Rv RTVG--EQQVVDEADRLIVNTDDEARQVISLHGADPARIDVVHPGVDLDV
MAV_4664|M.avium_104 RTVG--EQQVVDEADRLIVNTDDEARQLISIHHADPARIDVVHPGVDLDV
MAB_1976|M.abscessus_ATCC_1997 LPVARSALRQIGESTDVITYVSRYTRGRFASAFGPRAALEYLPSGVDTNR
TH_1111|M.thermoresistible__bu LPVARHALRIIGDNTDVLTYVSRYTRRRFASAFGPRAGLEHVPPGVDIDR
MLBr_00886|M.leprae_Br4923 LPVARSVLRRIGDVTDVVTFVSRYTRSRVTAAFGPRAALEYLPPGVDADR
Mflv_5216|M.gilvum_PYR-GCK RSVL---EEVLALAGCAVVMSEAARSLLCAGYAVDPQKVIIIPHGAAPAE
Mvan_1052|M.vanbaalenii_PYR-1 RSVL---EDVLALAGRAVVMSASARARLCAGYTVDPDKVSIIPHGAALPD
MSMEG_1207|M.smegmatis_MC2_155 RAVL---EQVAALAARVVVMSASASRRLCRDFRVDRRKVITIPHGATLPC
MUL_1124|M.ulcerans_Agy99 PVLG---RLGWFVESKLSPWLNRRNQYVTVSLPSARDLVALGVDNERIAV
MMAR_4514|M.marinum_M APPARAAVRRMLAADHWVVLGERHVQEYASRLRLPTSRISVLPNAVRIPN
:
Mb0496|M.bovis_AF2122/97 FRPGDRRAARAALGLPVDER-VVAFVGRIQPLKAPDIVLRAAAKLPGVRI
Rv0486|M.tuberculosis_H37Rv FRPGDRRAARAALGLPVDER-VVAFVGRIQPLKAPDIVLRAAAKLPGVRI
MAV_4664|M.avium_104 FRPGDRRAARAALGLPLDED-IVAFVGRIQPLKAPDIVLRAAAKLPGVRI
MAB_1976|M.abscessus_ATCC_1997 FCPDPAAREELRERYGLGDRPTIVCVSRLVPRKGQDMLIEALPAIR-ERV
TH_1111|M.thermoresistible__bu FAPDPVARTALRTRYRLGERPVVLCLSRLVPRKGQDMLIRALPMIR-RRV
MLBr_00886|M.leprae_Br4923 FRPDPASRAELRNRYRLGERPTVVCLARLVPRKGQDMLIRALPSIR-QRA
Mflv_5216|M.gilvum_PYR-GCK DVEWKPAVRPTILTWGLLGP--GKGVERVIDVMASLQDLPDPPRYLIAGR
Mvan_1052|M.vanbaalenii_PYR-1 AVAPQPVGRPTILTWGLLGP--GKGIERVIDAMVTLKDLPAAPRYLIAGR
MSMEG_1207|M.smegmatis_MC2_155 GPVLKRGGRPTLLTWGLLGP--GKGIERGIDAMSVLRRLNGMPRYLIAGL
MUL_1124|M.ulcerans_Agy99 VRNGLDEAPGQTLSGPRAATPRVVVLSRLVPHKQIEDALEAVAQLRRRTP
MMAR_4514|M.marinum_M TPVNQLG----------VQRVHAVALGRLGVRKGSYDLIDAVAALDETVR
: * : .
Mb0496|M.bovis_AF2122/97 IVAGGPS--GSGLASPDGLVRLADELGISARVTFLPPQ-SHTDLATLFRA
Rv0486|M.tuberculosis_H37Rv IVAGGPS--GSGLASPDGLVRLADELGISARVTFLPPQ-SHTDLATLFRA
MAV_4664|M.avium_104 VVAGGPS--GTGLASPDGLALLADELGISARVTFLPPQ-SRPNLATLFQA
MAB_1976|M.abscessus_ATCC_1997 DGAALVI--VGGGPYAEPLRALANRFGVDEHVVFTGGV-PWEELPAHHAM
TH_1111|M.thermoresistible__bu GDAVLVI--VGGGPYRTALQRLAHRYDVAAHVVFTGGI-PAEELPAHHAM
MLBr_00886|M.leprae_Br4923 DGAALMI--VGGGPYLGTLRKLAQGCGVADHVIFAGGV-AAVEVPAHYTL
Mflv_5216|M.gilvum_PYR-GCK THPKVLA--AQGEAYRDGLAARADRLGVGDSVIFDATYRSPQALAEMLRD
Mvan_1052|M.vanbaalenii_PYR-1 THPKVLA--THGEAYRDGLLAQARRLGVADSVVFDATYRTPQALTTLLQD
MSMEG_1207|M.smegmatis_MC2_155 THPKVLA--AEGEAYRDACVERARSTGVSDAVYFDPGYQSASSLTTLVQA
MUL_1124|M.ulcerans_Agy99 GSPNLHIDVVGGGWWRQRLIDHVHRLGISDAVTFHGHV-DDVTKHHVLQS
MMAR_4514|M.marinum_M NRLLATL---AGDGDVAQVRAAVAKAGLGETIDVVGWL-NPMARDELLRR
* . .: : .
Mb0496|M.bovis_AF2122/97 ADLVAVPSYS-------ESFGLVAVEAQACGTPVVAA-AVGGLPVAVRDG
Rv0486|M.tuberculosis_H37Rv ADLVAVPSYS-------ESFGLVAVEAQACGTPVVAA-AVGGLPVAVRDG
MAV_4664|M.avium_104 ANLVAVPSYS-------ESFGLVALEAQACGTPVAAA-AVGGLPVAVRDG
MAB_1976|M.abscessus_ATCC_1997 GDVFAMPCRTRGAGLDVEGLGIVFLEASACGVPVVAG-NSGGAPETVRAG
TH_1111|M.thermoresistible__bu ADVFAMPCRTRGAGLDVEGLGIVYLEASATGVPVVAG-RSGGAPETVRHG
MLBr_00886|M.leprae_Br4923 ADVFAMPCRTRGGGMDVEGLGIVFLEASATGVPVIAG-KSGGAPEAVQHN
Mflv_5216|M.gilvum_PYR-GCK AAVVVLPYDSKD-----QVTSGVLIDSIANGRPVIAT-AFPHAVEVLGNG
Mvan_1052|M.vanbaalenii_PYR-1 AAVVVLPYDSKE-----QVTSGVLVDSVASGRPVIAT-AFPHAVELLGTG
MSMEG_1207|M.smegmatis_MC2_155 AKVVLLPYDSTE-----QVTSGVLVDAIAHGRPVVAT-AFPHAVELLSSG
MUL_1124|M.ulcerans_Agy99 SWVQLLPSRK-------EGWGLAVVEAAQHGVPTIGYRSSGGLSDSIIDG
MMAR_4514|M.marinum_M AQIFVLPSRN-------EGLPMALLEAMANGLAPVTT-TAGSIGEVVTDG
. : :* . : . ::: * . : .
Mb0496|M.bovis_AF2122/97 ITGTLVSGHEVGQWADAIDHLLRLCAGPRGRVMSRAAARHAATFSWENTT
Rv0486|M.tuberculosis_H37Rv ITGTLVSGHEVGQWADAIDHLLRLCAGPRGRVMSRAAARHAATFSWENTT
MAV_4664|M.avium_104 VTGTLVAGHDVDHWADALAGLLR-APAPAAAAMSRAAAAHAATFSWDHTT
MAB_1976|M.abscessus_ATCC_1997 ETGVVVDGRSVSAITDAIVQILV---DPARAAAMGAAGRAWVTEQWRWDH
TH_1111|M.thermoresistible__bu ETGLVVDGRDVREIATAVGDLLS---DPDRAAAMGAAGRQWVVQQWQWQT
MLBr_00886|M.leprae_Br4923 KTGLVVDGRSVNMVADAVTELLT---DRDRAAAMGAAGRHWVTAEWRWDT
Mflv_5216|M.gilvum_PYR-GCK -AGTVVDHDDPEALTDALRRLIT--DPDTAGSMAAVARTLAPSVAWPVVA
Mvan_1052|M.vanbaalenii_PYR-1 -AGTVVDHDDAGALVDALRRLLT--DSRAAGSMAFAARELAPAMAWPVVA
MSMEG_1207|M.smegmatis_MC2_155 -AGIVVDHDDPDALVSALRSVIT--QPRLAGAMAAEARRIAPSMAWSVVA
MUL_1124|M.ulcerans_Agy99 VTGILVDSH--AELVDQLERLLA--DPVLRDQLGAKARVRSAEFSWQESV
MMAR_4514|M.marinum_M VNGLLVGPAHADQLAHALGALVT--DENLRARLGGAARERAGEFGLDRWY
* :* . : :: *
Mb0496|M.bovis_AF2122/97 DALLASYRRAIGEYNAERQRRGGEVISDLVAVGKPRHWTPRRGVGA
Rv0486|M.tuberculosis_H37Rv DALLASYRRAIGEYNAERQRRGGEVISDLVAVGKPRHWTPRRGVGA
MAV_4664|M.avium_104 DALLASYRRAIREYTTER-----------------------RGVGA
MAB_1976|M.abscessus_ATCC_1997 HAARFADLVSGSA---------------------------------
TH_1111|M.thermoresistible__bu QADRLARLLQV-----------------------------------
MLBr_00886|M.leprae_Br4923 LAARLADLLCGNDSR-------------------------------
Mflv_5216|M.gilvum_PYR-GCK DAYAALAQRLVAEKLATA----------------------------
Mvan_1052|M.vanbaalenii_PYR-1 GAYIALAQRLVAERQAAA----------------------------
MSMEG_1207|M.smegmatis_MC2_155 NAYQTVARELLTGRPRQR----------------------------
MUL_1124|M.ulcerans_Agy99 DGMRSVLEAVPAGRFVSGVV--------------------------
MMAR_4514|M.marinum_M QQLTRLWASLAGDPHPSRR---------------------------