For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MLSPMTTAETETGVLAGDERMLIDGELQNTGSGATFDVIHPASEQVAGVATDGTVDDMSRAVGAARRAFD TTDWSRDLDFRYHCITQLHEALERNKERLRRVLITEVGCPVSVTGSQIESPIEEVKHWAEHGRNFDYLVD NGVHDTPLGPARRKIHYEAVGVVGAITPWNVPFYLNIAETVPALMAGNTVVLKPAQLTPWSGSEYGRIVA EETDIPAGVFNVVVSNANEVGAALSSDPRVDMITFTGSTATGRAILAAGASTVKKTLLELGGKSAHIVLD DADFNSALSMAAMMACVMSGQSCILPSRILLPRSRYDEGIEILKNMMENFPVGDPWTPGIMQGPQISDTQ RQKVLGLIKSGIDSGARLVTGGGIPENLPVGYYTQPTLLADVDPDSQVAQEEIFGPVLTVTPYDTDDDAV AIANNSIYGLSGEVSGGDLERAFAVATRMRTGNVTINGKSHFGIGSPFGGTKQSGLGYRNGEEGYKEYLD AKTIGMPDQ
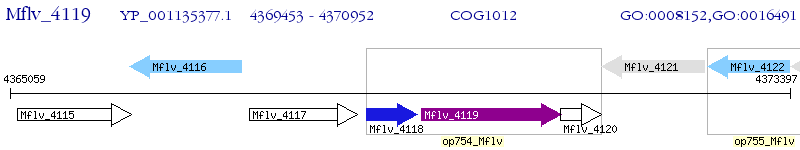
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4119 | - | - | 100% (499) | aldehyde dehydrogenase |
| M. gilvum PYR-GCK | Mflv_1600 | - | e-130 | 49.69% (479) | aldehyde dehydrogenase |
| M. gilvum PYR-GCK | Mflv_2502 | - | e-121 | 49.08% (491) | aldehyde dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0791 | aldA | 1e-131 | 49.58% (480) | aldehyde dehydrogenase NAD dependant AldA |
| M. tuberculosis H37Rv | Rv0768 | aldA | 1e-131 | 49.58% (480) | aldehyde dehydrogenase NAD dependent AldA |
| M. leprae Br4923 | MLBr_02573 | gabD1 | 1e-50 | 29.37% (463) | succinic semialdehyde dehydrogenase |
| M. abscessus ATCC 19977 | MAB_1218 | - | 1e-126 | 48.65% (483) | aldehyde dehydrogenase AldA |
| M. marinum M | MMAR_4928 | aldA | 1e-125 | 48.33% (480) | NAD-dependent aldehyde dehydrogenase, AldA |
| M. avium 104 | MAV_0713 | - | 1e-126 | 47.92% (480) | aldehyde dehydrogenase (NAD) family protein |
| M. smegmatis MC2 155 | MSMEG_2552 | - | 0.0 | 92.91% (494) | aldehyde dehydrogenase (NAD) family protein |
| M. thermoresistible (build 8) | TH_3081 | - | 0.0 | 83.84% (495) | PUTATIVE aldehyde dehydrogenase |
| M. ulcerans Agy99 | MUL_0477 | aldA | 1e-125 | 48.33% (480) | NAD-dependent aldehyde dehydrogenase, AldA |
| M. vanbaalenii PYR-1 | Mvan_2226 | - | 0.0 | 93.54% (495) | aldehyde dehydrogenase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4119|M.gilvum_PYR-GCK MLSPMTTAETETGVLAGDERMLIDGELQNTGSGATFDVIHPASEQVAGVA
Mvan_2226|M.vanbaalenii_PYR-1 ----MTTTETDTGVLAGDERMLIDGELQITGSGATFDVIHPASEQVAGRA
MSMEG_2552|M.smegmatis_MC2_155 ----MTSVQDETGVLAGDERMLIDGELQTTGSGSTFDVIHPATEQVAGQA
TH_3081|M.thermoresistible__bu ----MTSVQDETGVLAGDERMLIDGELQYTSSGAKFDVIHPANEEVAGQA
Mb0791|M.bovis_AF2122/97 ----MALWGD------GISALLIDGKLSDGRAG-TFPTVNPATEEVLGVA
Rv0768|M.tuberculosis_H37Rv ----MALWGD------GISALLIDGKLSDGRAG-TFPTVNPATEEVLGVA
MMAR_4928|M.marinum_M ----MALLAD------GVSSLFIDGKRCEGAAG-TFATVNPATEETLGVA
MUL_0477|M.ulcerans_Agy99 ----MALLAD------GVSSLFIDGKRCEGAAG-TFATVNPATEETLGVA
MAV_0713|M.avium_104 ----MALLAD------GVSELFIDGKMSAGNAG-TFPTVNPATEEVLGVA
MAB_1218|M.abscessus_ATCC_1997 ----MTVLSD------GDSELFIDGKLLPGGAG-RFPTVNPATEEVLGTA
MLBr_02573|M.leprae_Br4923 ---------------------------------MSIATINPATGETVKTF
: .::**. :.
Mflv_4119|M.gilvum_PYR-GCK TDGTVDDMSRAVGAARRAFDTTDWSRDLDFRYHCITQLHEALERNKERLR
Mvan_2226|M.vanbaalenii_PYR-1 TDGTVEDMARACSAARRAFDETDWSRDLDFRYHCLTQLHEALERNKERLR
MSMEG_2552|M.smegmatis_MC2_155 TDGTVEDMSRAVAAARRAFDQTDWSRDLDFRYHCLTQLHEALERNKERLR
TH_3081|M.thermoresistible__bu TDGTVADMERAVAAARRAFDTTDWSRDVEFRHHCLIQLAEALEAEKERLR
Mb0791|M.bovis_AF2122/97 ADADAEDMGRAIEAARRAFDSTDWSRNTELRVRCVRQLRDAMQQHVEELR
Rv0768|M.tuberculosis_H37Rv ADADAEDMGRAIEAARRAFDSTDWSRNTELRVRCVRQLRDAMQQHVEELR
MMAR_4928|M.marinum_M ADADAGDMDRAIEAARRAFDDTDWSRNTELRVRCVRQLRDAMREHIEELR
MUL_0477|M.ulcerans_Agy99 ADAVAGDMDRAIEAARRAFDDTDWSRNTELRVRCVRQLRDAMREHIEELR
MAV_0713|M.avium_104 ANADAADMDRAIDAARRAFDETDWSRNTELRVRCVRQLRDAMREHIEELR
MAB_1218|M.abscessus_ATCC_1997 ADADSDDMSDAIEAARRAFDDSGWSRDVALRVHCIRQLRDGMKTHIEELR
MLBr_02573|M.leprae_Br4923 TPTTQDEVEDAIARAYARFDN-YRHTTFTQRAKWANATADLLEAEANQSA
: :: * * ** * : : :. . :.
Mflv_4119|M.gilvum_PYR-GCK RVLITEVGCP-VSVTGSQIESPIEEVKHWAEHGRNFDYLVDNGVHDTPLG
Mvan_2226|M.vanbaalenii_PYR-1 RILITEVGCP-VSVSGSQIESPIEEVKHWAEHGRDFDYLVDNGVHDTPLG
MSMEG_2552|M.smegmatis_MC2_155 RILITEVGCP-VTVSGSQIESPIEEVRHWAEHGRNFDYLVDTGVHPTQLG
TH_3081|M.thermoresistible__bu RILVTEVGCP-VTVTGSQIDEPINEVRHWAEHGKAFQYIVDNGVHDTPLG
Mb0791|M.bovis_AF2122/97 ELTISEVGAPRMLTASAQLEGPVGDLSFAADTAESYPWKQDLGEASPLGI
Rv0768|M.tuberculosis_H37Rv ELTISEVGAPRMLTASAQLEGPVGDLSFAADTAESYPWKQDLGEASPLGI
MMAR_4928|M.marinum_M ALTISEVGAPRMLTAAAQLEGPVNDLAFSADTAESYSWKQDLGAAAPMGI
MUL_0477|M.ulcerans_Agy99 ALTISEVGAPRMLTAAAQLEGPVNDLAFSADTAESYSWKQDLGAAAPMGI
MAV_0713|M.avium_104 DITIAEVGAPRMLTSAAQLEGPVGDLSFAADTAESYEWNQDLGQASPMGI
MAB_1218|M.abscessus_ATCC_1997 ELTIAEVGAPRMLTAGAQLEGPVDDLQFAADTAESFDWTSDLGVASPMGI
MLBr_02573|M.leprae_Br4923 ALMTLEMGKT-LQSAKDEAMKCAKGFRYYAEKAET--LLADEPAEAAAVG
: *:* . : : : . . *: .. * .
Mflv_4119|M.gilvum_PYR-GCK PARRKIHYEAVGVVGAITPWNVPFYLNIAETVPALMAGNTVVLKPAQLTP
Mvan_2226|M.vanbaalenii_PYR-1 PARRKIHYEAVGVVGAITPWNVPFYLNIAETVPALMAGNTVVLKPAQLTP
MSMEG_2552|M.smegmatis_MC2_155 PARRKIHYEPVGVVGAITPWNVPFYLNIAETVPALMAGNTVVLKPAQLTP
TH_3081|M.thermoresistible__bu PAHRKIVYEATGVVGAITPWNVPFYLNIAETMPALMAGNTVVLKPAQLTP
Mb0791|M.bovis_AF2122/97 ATRRTLAREAVGVVGAITPWNFPHQINLAKLGPALAAGNTVVLKPAPDTP
Rv0768|M.tuberculosis_H37Rv ATRRTLAREAVGVVGAITPWNFPHQINLAKLGPALAAGNTVVLKPAPDTP
MMAR_4928|M.marinum_M PTRRTVVREAVGVVGAITPWNFPHQINLAKLGPALAAGNTVVLKPAPDTP
MUL_0477|M.ulcerans_Agy99 PTRRTVVREAVGVVGAITPWNFPHQINLAKLGPALAAGNTVVLKPAPDTP
MAV_0713|M.avium_104 PTRRTIAREAVGVVGAITPWNFPHQINLAKLGPALAAGNTVVLKPAPDTP
MAB_1218|M.abscessus_ATCC_1997 KTRRTLAREAVGVVGAVTPWNFPHQINLAKVGPALAAGNTLVLKPAPDTP
MLBr_02573|M.leprae_Br4923 ASKALARYQPLGVVLAVMPWNFPLWQTVRFAAPALMAGNVGLLKHASNVP
:: :. *** *: ***.* .: *** ***. :** * .*
Mflv_4119|M.gilvum_PYR-GCK WSGSEYGRIVAEETDIPAGVFNVVVSNANEVGAALSSDPRVDMITFTGST
Mvan_2226|M.vanbaalenii_PYR-1 WSGTEYGRIVAEETDIPAGVFNVVVSNANEVGAALSADPRVDMITFTGST
MSMEG_2552|M.smegmatis_MC2_155 WSGTEYGRIVAEETDIPAGVFNVVVSNANEVGAALSADPRVDMITFTGST
TH_3081|M.thermoresistible__bu WSGTELGRIVAEHTDIPPGVFNVVVSNANEVGAALTADPRVDMITFTGST
Mb0791|M.bovis_AF2122/97 WCAAALGEIIVEHTDFPPGVVNIVTSSSHALGALLAKDPRVDMISFTGST
Rv0768|M.tuberculosis_H37Rv WCAAALGEIIVEHTDFPPGVVNIVTSSSHALGALLAKDPRVDMISFTGST
MMAR_4928|M.marinum_M WCAAVLGELIADCTDIPPGVVNIVTSSEHGLGALLAKDPRVDMVSFTGST
MUL_0477|M.ulcerans_Agy99 WCAAVLGELIADCTDIPPGVVNIVTSSEHGLGALLAKDPRVDMVSFTGST
MAV_0713|M.avium_104 WCAAVLGQLIAEHTDFPPGVVNIITSDDHGVGALLSKDPRVDMVSFTGST
MAB_1218|M.abscessus_ATCC_1997 WCAAVLGRIIAEHTDFPPGVVNIVTSSDHRIGAQLSTDPRVDMVSFTGST
MLBr_02573|M.leprae_Br4923 QCALYLADVIARG-GFPEDCFQTLLISANGVEAILR-DPRVAAATLTGSE
.. . ::. .:* . .: : . : : * * **** ::***
Mflv_4119|M.gilvum_PYR-GCK ATGRAILAAGASTVKKTLLELGGKSAHIVLDD---ADFNSALSMAAMMAC
Mvan_2226|M.vanbaalenii_PYR-1 ATGRAILAAGAPTVKKTLLELGGKSAHIVLDD---ADFNAALPLAAMMAC
MSMEG_2552|M.smegmatis_MC2_155 ATGRAILAAGAPTVKKTLLELGGKSAHIVLDD---ADFNSALSMAAMMAC
TH_3081|M.thermoresistible__bu ATGRAILAAAAPTVKKTMLELGGKSAHIVLDD---ADLNASLPLAAMMAC
Mb0791|M.bovis_AF2122/97 ATGRAVMADAAATIKKVFLELGGKSAFVVLDD---ADLAAASAVSAFSAC
Rv0768|M.tuberculosis_H37Rv ATGRAVMADAAATIKKVFLELGGKSAFVVLDD---ADLAAASAVSAFSAC
MMAR_4928|M.marinum_M ATGRSVMADGAATIKRMFLELGGKSAFIVLDD---ADLAAASSVSAFTAS
MUL_0477|M.ulcerans_Agy99 ATGRSVMADGAATIKRMFLELGGKSAFIVLDD---ADLAAASSVSAFTAS
MAV_0713|M.avium_104 VTGRSVMSDAAATIKRVFLELGGKSAFVVLDD---ADLGGACSMSAFTAA
MAB_1218|M.abscessus_ATCC_1997 ATGRAVMTDAAATVKKVFLELGGKSAFIVLDDLDEAALGSACSVAAFTAA
MLBr_02573|M.leprae_Br4923 PAGQAVGAIAGNEIKPTVLELGGSDPFIVMPS---ADLDAAVATAVTGRV
:*::: : .. :* .*****....:*: . * : .: . :.
Mflv_4119|M.gilvum_PYR-GCK VMSGQSCILPSRILLPRSRYDEGIEILKNMMENFPVGDPWTPGIMQGPQI
Mvan_2226|M.vanbaalenii_PYR-1 VMSGQSCILPSRILLPRSRYDEGIEILKTMMEGFPVGDPWTPGNMQGPQI
MSMEG_2552|M.smegmatis_MC2_155 VMSGQSCILPSRILLPRSRYDEGIEILKNMMENFPVGDPWTPGVMQGPQI
TH_3081|M.thermoresistible__bu VMSGQSCILPSRILLPRSRYDEGLEILKGAMANFPMGDPWTPGNMQGPQI
Mb0791|M.bovis_AF2122/97 MHAGQGCAITTRLVVPRARYEEAVAIAAATMSSIRPGDPNDPGTVCGPLI
Rv0768|M.tuberculosis_H37Rv MHAGQGCAITTRLVVPRARYEEAVAIAAATMSSIRPGDPNDPGTVCGPLI
MMAR_4928|M.marinum_M MHAGQGCAITTRLVVPRARYDEAVAIAAGTMSSIKPGDPDDARTVCGPLI
MUL_0477|M.ulcerans_Agy99 MHAGQGCAITTRLVVPRARYDEAVAIAAGTMSSIKPGDPDDARTVCGPLI
MAV_0713|M.avium_104 MHAGQGCAITTRLLVPRDRYDEAVAAAAGTMGSIKPGDPNDPGTVCGPVI
MAB_1218|M.abscessus_ATCC_1997 MHAGQGCAITTRLVVPRERYDDAVAAAAATMGSLKPGDPAKPGTICGPVI
MLBr_02573|M.leprae_Br4923 QNNGQSCIAAKRFIAHADIYDEFVEKFVARMETLRVGDPTDPDTDVGPLA
**.* ..*:: *:: : * : *** . **
Mflv_4119|M.gilvum_PYR-GCK SDTQRQKVLGLIKSGIDSGARLVTGGGIPENLPVGYYTQPTLLADVDPDS
Mvan_2226|M.vanbaalenii_PYR-1 SEAQRQKVLGLIKSGIDSGARLVTGGGIPENLPVGYYTQPTLLADVDPDS
MSMEG_2552|M.smegmatis_MC2_155 SETQRQKVLGLIRSGIDSGARLVTGGGVPENLPVGYYTQATLLADVDPDS
TH_3081|M.thermoresistible__bu SEAQRQKVLGLIKSGLDSGARLVLGGGIPENLPVGYYTQPTLLADVDPNS
Mb0791|M.bovis_AF2122/97 SARQRDRVQGYLDLAVAEGGRFACGGARPADREVGFYIEPTVIAGLTNDA
Rv0768|M.tuberculosis_H37Rv SARQRDRVQGYLDLAVAEGGRFACGGARPADREVGFYIEPTVIAGLTNDA
MMAR_4928|M.marinum_M SQRQRDRVQGYLDLAIAEGGTFACGGGRPAGRQVGYFIEPTVIAGLTNDA
MUL_0477|M.ulcerans_Agy99 SQRQRDRVEGYLDLAIAEGGTFACGGGRPAGRQVGYFIEPTVIAGLTTDA
MAV_0713|M.avium_104 SARQRDRVQGYLDLAIAEGGTFACGGGRPADKDVGFFIEPTVIAGLTNDA
MAB_1218|M.abscessus_ATCC_1997 SARQRDRVQSYLDLAVAEGGSFACGGGRPEGKDRGFFIEPTVVAGLTNAA
MLBr_02573|M.leprae_Br4923 TESGRAQVEQQVDAAAAAGAVIRCGGKRPDRP--GWFYPPTVITDITKDM
: * :* : . *. : ** * *:: .*:::.:
Mflv_4119|M.gilvum_PYR-GCK QVAQEEIFGPVLTVTPYDTDDDAVAIANNSIYGLSGEVSGGDLERAFAVA
Mvan_2226|M.vanbaalenii_PYR-1 QVAQEEIFGPVLTVTPYDTDDDAIAIANNTIYGLSGEVSSADVDRAFAVA
MSMEG_2552|M.smegmatis_MC2_155 QVAQEEIFGPVLTVTPYDTDDDAVAIANNTIYGLSGEVSGGDLDRAFAIA
TH_3081|M.thermoresistible__bu QVAQEEIFGPVLTVTPYDTEDEAIEIANNTIYGLSGEVSSADVDRAFRVA
Mb0791|M.bovis_AF2122/97 RVAREEIFGPVLTVIAHDGDDDAVRIANDSPYGLSGTVYGADPQRAARIA
Rv0768|M.tuberculosis_H37Rv RVAREEIFGPVLTVIAHDGDDDAVRIANDSPYGLSGTVYGADPQRAARIA
MMAR_4928|M.marinum_M RPAREEIFGPVLTVLAHDGDDDAVRIANDSPYGLSGTVYGGDPQRAADVA
MUL_0477|M.ulcerans_Agy99 RPAREEIFGPVLTVLAHDGDDDAVRIANDSPYGLSGTVYGGDPQRAADVA
MAV_0713|M.avium_104 RPAREEIFGPVLTVIAYDGEEDALRIANDSPYGLSGTVFSADPERAANFA
MAB_1218|M.abscessus_ATCC_1997 RVAREEIFGPVLVVIAHDGDEDAVRIANDSPYGLSGTVFGDDDERSARVA
MLBr_02573|M.leprae_Br4923 ALYTDEVFGPVASVYCATNIDDAIEIANATPFGLGSNAWTQNESEQRHFI
:*:**** * ::*: *** : :**.. . : .. .
Mflv_4119|M.gilvum_PYR-GCK TRMRTGNVTINGKSHFGIGSPFGGTKQSGLGYRNGEEGYKEYLDAKTIGM
Mvan_2226|M.vanbaalenii_PYR-1 CRMRTGNVTINGKSHFGIGSPFGGTKQSGLGYRNGEEGYKEYLEAKTIGM
MSMEG_2552|M.smegmatis_MC2_155 CRMRTGNVTINGKSHFGITSPFGGTKQSGLGYRNGEEGYKEYLEAKTIGM
TH_3081|M.thermoresistible__bu QRMRTGNVTINGRSHFGINSPFGGTKQSGLGRRNGEEGFKEYLESKTVGM
Mb0791|M.bovis_AF2122/97 SRLRVGTVNVNGGVWYCADAPFGGYKQSGIGREMGLLGFEEYLEAKLIAT
Rv0768|M.tuberculosis_H37Rv SRLRVGTVNVNGGVWYCADAPFGGYKQSGIGREMGLLGFEEYLEAKLIAT
MMAR_4928|M.marinum_M ARLRVGTVNVNGGVWYCADAPFGGYKQSGIGREMGLAGFEEYLETKLIAT
MUL_0477|M.ulcerans_Agy99 ARLRVGAVNVNGGVWYCADAPFGGYKQSGIGREMGLAGFEEYLETKLIAT
MAV_0713|M.avium_104 SRMRVGTVNVNGGVWYSADAPFGGYKQSGNGREMGLAGFEEYLEIKTIAT
MAB_1218|M.abscessus_ATCC_1997 SRLRVGTVNVNGGVWYSADAPFGGYKQSGNGREMGVAGFEEYLETKVIAT
MLBr_02573|M.leprae_Br4923 DNIVAGQVFINGMTVSYPELPFGGIKRSGYGRELSTHGIREFCNIKTVWI
.: .* * :** **** *:** * . . * .*: : * :
Mflv_4119|M.gilvum_PYR-GCK PDQ-
Mvan_2226|M.vanbaalenii_PYR-1 PDQ-
MSMEG_2552|M.smegmatis_MC2_155 PE--
TH_3081|M.thermoresistible__bu PPQE
Mb0791|M.bovis_AF2122/97 AAN-
Rv0768|M.tuberculosis_H37Rv AAN-
MMAR_4928|M.marinum_M AAN-
MUL_0477|M.ulcerans_Agy99 AAN-
MAV_0713|M.avium_104 AVQ-
MAB_1218|M.abscessus_ATCC_1997 AVK-
MLBr_02573|M.leprae_Br4923 A---
.