For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTARTPNLRPVRELTPTLRYCTIHGYRRAYRIAGSGPAILLIHGIGDNSTTWAGVQTQLAQRFTVIAPDL LGHGKSDKPRADYSVAAYANGMRDLLSVLNIDSATVVGHSLGGGVAMQFAYQFPQLVDRLILVGAGGVTK DVNIALRAASLPMGTEALALLRLPLLLPTLRLLGRVGGPLLGNTRIGRDIPNMLRILADLPEPTASSAFA RTLRSVVDWRGQVVTMLDRCYLTQSVPVQLIWGSQDSVIPVSHAEMAHTAMPGSRLEIFEGAGHFPFHDE PDRFVELVEKFIDSTEPAVYDQEYLRTLLRTGTSENTLSGDVDTQVAVLDAIGADERSAT
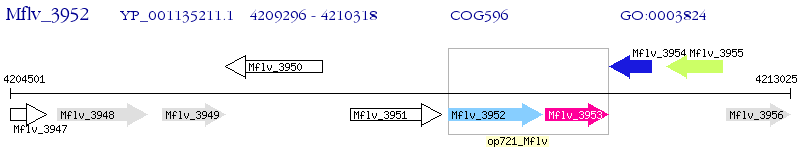
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3952 | - | - | 100% (340) | alpha/beta hydrolase fold |
| M. gilvum PYR-GCK | Mflv_2367 | - | 3e-63 | 47.37% (285) | alpha/beta hydrolase fold |
| M. gilvum PYR-GCK | Mflv_2400 | - | 2e-20 | 25.71% (280) | alpha/beta hydrolase fold |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2734 | - | 1e-150 | 74.49% (341) | hydrolase |
| M. tuberculosis H37Rv | Rv2715 | - | 1e-150 | 74.49% (341) | hydrolase |
| M. leprae Br4923 | MLBr_00685 | - | 7e-11 | 27.87% (287) | putative hydrolase |
| M. abscessus ATCC 19977 | MAB_3034 | - | 1e-135 | 71.69% (325) | hydrolase |
| M. marinum M | MMAR_1997 | - | 1e-154 | 75.95% (341) | hydrolase |
| M. avium 104 | MAV_3609 | - | 1e-149 | 75.15% (338) | hydrolase |
| M. smegmatis MC2 155 | MSMEG_4867 | - | 2e-19 | 26.37% (273) | 2-hydroxy-6-ketonona-2,4-dienedoic acid hydrolase |
| M. thermoresistible (build 8) | TH_4528 | - | 6e-21 | 25.89% (282) | PUTATIVE alpha/beta hydrolase fold |
| M. ulcerans Agy99 | MUL_3358 | - | 1e-153 | 75.66% (341) | hydrolase |
| M. vanbaalenii PYR-1 | Mvan_2445 | - | 1e-170 | 85.29% (340) | alpha/beta hydrolase fold |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3952|M.gilvum_PYR-GCK MTARTPN--LRPVR------------------ELT-PTLRYCTIHG----
Mvan_2445|M.vanbaalenii_PYR-1 MTERKPN--LRPVR------------------ELT-PTLKFCTIHG----
Mb2734|M.bovis_AF2122/97 MTERKRN--LRPVR------------------DVAPPTLQFRTVHG----
Rv2715|M.tuberculosis_H37Rv MTERKRN--LRPVR------------------DVAPPTLQFRTVHG----
MMAR_1997|M.marinum_M MTERKRN--LRPVR------------------EVTAPSLQFRTIHG----
MUL_3358|M.ulcerans_Agy99 MTERKRN--LRPVR------------------EVTAPSLQFRTIHG----
MAV_3609|M.avium_104 MTERKRKNTLRPVR------------------EVSAPRLEFRTIHG----
MAB_3034|M.abscessus_ATCC_1997 MNPPPRL--LRPVPDTEHIDGVDGGGDTPSAATSTRPRLAFRTIHG----
MSMEG_4867|M.smegmatis_MC2_155 MTSTAAAGTASAVEFES----------IWSDLQGVAFSQGYLDVPVGDRK
TH_4528|M.thermoresistible__bu ---------------------------VWSDLQGVAFTQGYLDAGG----
MLBr_00685|M.leprae_Br4923 MPACQHQR-HRPVV-------------------ACGTRGTTFRVGS----
Mflv_3952|M.gilvum_PYR-GCK YRRAYRIAGS--GPAILLIHGIGDNSTTWAGVQTQLA--QRFTVIAPDLL
Mvan_2445|M.vanbaalenii_PYR-1 YRRAFRVAGS--GPAILLIHGIGDNSTTWSTVQTQLA--QRFTVIAPDLL
Mb2734|M.bovis_AF2122/97 YRRAFRIAGS--GPAILLIHGIGDNSTTWNGVHAKLA--QRFTVIAPDLL
Rv2715|M.tuberculosis_H37Rv YRRAFRIAGS--GPAILLIHGIGDNSTTWNGVHAKLA--QRFTVIAPDLL
MMAR_1997|M.marinum_M YKRAFRIAGS--GPAILLIHGIGDNSTTWTGIHAKLA--QRFTVIAPDLL
MUL_3358|M.ulcerans_Agy99 YKRAFRIAGS--GPAILLIHGIGDNSTTWTGIHAKLA--QRFTVIAPDLL
MAV_3609|M.avium_104 YRRAYRIAGS--GPAILLIHGIGDNSTTWNTVQAKLA--QRFTVIAPDLL
MAB_3034|M.abscessus_ATCC_1997 YRRAYRIAGS--GPVLLLIHGIGDNSATWDSVHAQLA--EHFTVIAPDLL
MSMEG_4867|M.smegmatis_MC2_155 IRTRYLHAGNPQAPALLFLHGSGGHAEAYVRNLAAHA--EHFSTWSIDML
TH_4528|M.thermoresistible__bu VRTRYLHAGEADKPVLIFLHGSGGHAEAYVRNLEAHA--EHFSTWSIDML
MLBr_00685|M.leprae_Br4923 -MRQPIHLGS--GEPVLLLHPFMMSQTVWEIVAPQLANTERYEVFAPTMA
*. ::::* .: * ::: . : :
Mflv_3952|M.gilvum_PYR-GCK G-HGKSDKPRADYSVAAYANGMRDLLSVLNIDSATVVGHSLGGGVAMQFA
Mvan_2445|M.vanbaalenii_PYR-1 G-HGRSDKPRADYSVAAYANGMRDLLSVLDIDDVTVIGHSLGGGVAMQFA
Mb2734|M.bovis_AF2122/97 G-HGQSDKPRADYSVAAYANGMRDLLSVLDIERVTIVGHSLGGGVAMQFA
Rv2715|M.tuberculosis_H37Rv G-HGQSDKPRADYSVAAYANGMRDLLSVLDIERVTIVGHSLGGGVAMQFA
MMAR_1997|M.marinum_M G-HGRSDKPRADYSIAAYANGMRDLLSVLDIERVTIIGHSLGGGVAMQFA
MUL_3358|M.ulcerans_Agy99 G-HGRSDKPRADYSIAAYANGMRDLLSVLDIERVTIIGHSLGGGVAMQFA
MAV_3609|M.avium_104 G-HGQSDKPRADYSVAAYANGMRDLLAVLDIERVTIVGHSLGGGVAMQFA
MAB_3034|M.abscessus_ATCC_1997 G-HGQSDKPRADYSVAAYANGMRDLLAVLDIERVTVVGHSLGGGVAMQFT
MSMEG_4867|M.smegmatis_MC2_155 G-HGYTDKPGHPLEVAHYVDHVIAFLDTIGAQRAHISGESLGGWVAARVA
TH_4528|M.thermoresistible__bu G-HGYTDKPGHPLEVRHYVEHLAAVLDAIGADRAHISGESLGGWVASRFA
MLBr_00685|M.leprae_Br4923 AHHGGPSASSWMMSVSTLADHVEQQLDELGWDTAHLVGNSLGGWVVFELE
. ** .. . .: .: : * :. : . : *.**** *. ..
Mflv_3952|M.gilvum_PYR-GCK YQFPQLVDRLILVGAGGVTKDVNIALRAASLPMGTEALALLR-LPLLLPT
Mvan_2445|M.vanbaalenii_PYR-1 YQFPQLVNRLILVGAGGVTKDVNIALRLASLPMGSEALALLR-LPLVLPA
Mb2734|M.bovis_AF2122/97 YQFPQLVDRLILVSAGGVTKDVNIVFRLASLPMGSEAMALLR-LPLVLPA
Rv2715|M.tuberculosis_H37Rv YQFPQLVDRLILVSAGGVTKDVNIVFRLASLPMGSEAMALLR-LPLVLPA
MMAR_1997|M.marinum_M YQFPQLVDRLILVGAGGVTKDVNVVFRLASLPMGAEALALLR-LPMVLPT
MUL_3358|M.ulcerans_Agy99 YQFPQLVDRLILVGAGGVTKDVNVVFRLASLPMGAEALALLR-LPMVLPT
MAV_3609|M.avium_104 YQFPHLVERLILVGAGGVTKDVNFVLRWASLPLGSEAIALLR-LPLVLPA
MAB_3034|M.abscessus_ATCC_1997 YQFPHLVERLILVAPGGVTKDVNIVLRCASLPFIGDALGLLR-LPMAMPM
MSMEG_4867|M.smegmatis_MC2_155 VDHPDRVEKLVLNTAGGSQADPEVMKRIITLSMAAAENPTWD-TVQARIK
TH_4528|M.thermoresistible__bu IDRPDRLDRLVLNTAGGSQADPEVMKRIITLSMAAAENPSWD-TVQARIT
MLBr_00685|M.leprae_Br4923 RRGRARSVTGIAPAGGWTRWSLAKFEVIAKFVAGMPLVAVARGLGSRALS
: * . .:
Mflv_3952|M.gilvum_PYR-GCK LRLLGRVGG--PLLGNTRIGRDIPNMLRILADLPEPTASSAFARTLRSVV
Mvan_2445|M.vanbaalenii_PYR-1 LQVLGRVSG--TVFGSTGVGRDIPDMLRILADLPEPTASSAFARTLRAVV
Mb2734|M.bovis_AF2122/97 VQIAGRIVG--KAIGTTSLGHDLPNVLRILDDLPEPTASAAFGRTLRAVV
Rv2715|M.tuberculosis_H37Rv VQIAGRIVG--KAIGTTSLGHDLPNVLRILDDLPEPTASAAFGRTLRAVV
MMAR_1997|M.marinum_M VQLAGKVLG--MALGSTALGQDLPNVLRILDDLPEPTASSAFTRTLRAVV
MUL_3358|M.ulcerans_Agy99 VQLAGKVLG--MALGSTALGQDLPNVLRILDDLPEPTASSAFTRTLRAVV
MAV_3609|M.avium_104 VQLMGRVLG--TALGSTGLGRDLPNVLRILDDLPEPTASAAFSRTLRAVV
MAB_3034|M.abscessus_ATCC_1997 LRLGGAVAR--ATFGRASMARDIPDVLRVLADLPEPRASAAFTRTLRAVV
MSMEG_4867|M.smegmatis_MC2_155 WLMADKSKDYDDIVASRQRVYRQPGFVAAMSDIMALQDPQIRARNLLGQA
TH_4528|M.thermoresistible__bu WLMADKSKGYDDIVASRQKIYRQPGFVDAMRNIMALQDPEIRARNLLGPE
MLBr_00685|M.leprae_Br4923 LPFSHRLAS--LPLTGSPAGLSEHQLRNIVEDAG--HCPAYFLLLIKTLL
. . . : : . :
Mflv_3952|M.gilvum_PYR-GCK DWRGQVVTMLDRCYLTQSVPVQLIWGSQDSVIP-VSHAEMAHTAMPG-SR
Mvan_2445|M.vanbaalenii_PYR-1 DWRGQVVTMLDRCYLTQSVPVQLIWGDCDAVIP-VSHGEMAHAAMPG-SR
Mb2734|M.bovis_AF2122/97 DWRGQMVTMLDRCYLTEAIPVQIIWGTKDVVLP-VRHAHMAHAAMPG-SQ
Rv2715|M.tuberculosis_H37Rv DWRGQMVTMLDRCYLTEAIPVQIIWGTKDVVLP-VRHAHMAHAAMPG-SQ
MMAR_1997|M.marinum_M DWRGQIVTMLDRCYLTQAIPVQIIWGSRDAVVP-VRHAEMAHAAMPG-SK
MUL_3358|M.ulcerans_Agy99 DWRGQIVTMLDRCYLTQAIPVQIIWGSRDAVVP-VRHAEMAHAAMPG-SK
MAV_3609|M.avium_104 DWRGQIVTMLDRCYLTEAIPVQIVWGTKDVVVP-VRHAWMAHAAMPG-SR
MAB_3034|M.abscessus_ATCC_1997 DWRGQVVTMLDRCYLTESVPVQLIWGSDDLVIP-VSHGHLAHAAMPG-SA
MSMEG_4867|M.smegmatis_MC2_155 EYG------------SITAPTLVVWTSDDPTAD-VAEGRRIASMIPG-AR
TH_4528|M.thermoresistible__bu EYG------------AITAPTLVVWTSDDPTAD-VDEGKRIASMIPG-AR
MLBr_00685|M.leprae_Br4923 QPG-------LRELADITVPVLLVLCEKDRVLPPPKYSRHFTDHLPERTQ
: : *. :: * . . :* :
Mflv_3952|M.gilvum_PYR-GCK LEIFEGAGHFPFHDEPDRFVELVEKFIDSTEPAVYDQEYLRTLLRTGTSE
Mvan_2445|M.vanbaalenii_PYR-1 LEVFEGSGHFPFHDDPDRFVEVVEKFIDTSEPAVYDHEYLRGLLRAGISE
Mb2734|M.bovis_AF2122/97 LEIFEGSGHFPFHDDPARFIDIVERFMDTTEPAEYDQAALRALLRRGGGE
Rv2715|M.tuberculosis_H37Rv LEIFEGSGHFPFHDDPARFIDIVERFMDTTEPAEYDQAALRALLRRGGGE
MMAR_1997|M.marinum_M LEVFEGSGHFPFHDDPARFIDIVLRFIDSTQPPEYDQAALRELLRTGGGE
MUL_3358|M.ulcerans_Agy99 LEVFEGSGHFPFHDDPARFIDIVLRFIDSTQPPEYDQAALRELLRTGGGE
MAV_3609|M.avium_104 LEIFEGSGHFPFHDDPARFIDVVERFIDSTEPAAYDQEALRRLLRSGGGA
MAB_3034|M.abscessus_ATCC_1997 LEIFDKSGHFPFHDDPERFIGIVRQFIASTEPGEYNEDELRRLLRTGVTE
MSMEG_4867|M.smegmatis_MC2_155 FEVMPGCGHWPQYEDPATFNALHLDFLLGR--------------------
TH_4528|M.thermoresistible__bu FEVMPGCGHWPQYEDPATFNRLHIDFLLGRS-------------------
MLBr_00685|M.leprae_Br4923 VTKFDGMGHVPMFEAPEVITKLITNFINQCSKPAEAASPAS---------
. : ** * .: * : : *:
Mflv_3952|M.gilvum_PYR-GCK NTLSGDVDTQVAVLDAIGADERSAT
Mvan_2445|M.vanbaalenii_PYR-1 GTISGPADTRVAVLDAMGADERSAT
Mb2734|M.bovis_AF2122/97 ATVTGSADTRVAVLNAIGSNERSAT
Rv2715|M.tuberculosis_H37Rv ATVTGSADTRVAVLNAIGSNERSAT
MMAR_1997|M.marinum_M RTVSGPADTRVAVLSAMGSDERSAT
MUL_3358|M.ulcerans_Agy99 RTVSDPADTRVAVLSAMGSDERSAT
MAV_3609|M.avium_104 QAVTGPAPTRVAVLSAMGSDERSAT
MAB_3034|M.abscessus_ATCC_1997 HAISGAARTRMAVLDAMGTDERSAT
MSMEG_4867|M.smegmatis_MC2_155 -------------------------
TH_4528|M.thermoresistible__bu -------------------------
MLBr_00685|M.leprae_Br4923 -------------------------