For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSTMDHPQGSPSEHSRGYRALVVDDEVPLAEVVASYLQREQFDAVVADNGVDAIALARDFDPDVVILDLG LPGIDGLEVCRSLRTFSDAYVVMLTARDTELDTVLGLTVGADDYVTKPFSPRELVARIRAMLRRPRLLQS PPTGADREVTPPRRFGALSIDVAAREVRIADDPILLTRTEFDILQALSGRPGIVLSRRQLLEIIRDEPWV GNDQLVDVHIGHLRRKLGDDAARPRYIATVRGVGYRMGTGA
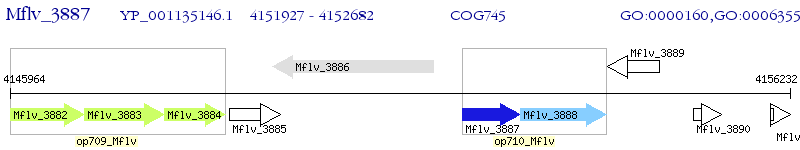
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3887 | - | - | 100% (251) | two component transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_2838 | - | e-120 | 85.20% (250) | two component transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_0893 | - | e-120 | 85.20% (250) | two component transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3274c | mtrA | 7e-45 | 43.91% (230) | two component sensory transduction transcriptional |
| M. tuberculosis H37Rv | Rv3246c | mtrA | 7e-45 | 43.91% (230) | two component sensory transduction transcriptional |
| M. leprae Br4923 | MLBr_00773 | mtrA | 4e-45 | 43.91% (230) | putative two-component response regulator |
| M. abscessus ATCC 19977 | MAB_3339 | - | 4e-94 | 69.66% (234) | putative sensory transduction protein |
| M. marinum M | MMAR_1299 | mtrA | 1e-44 | 43.91% (230) | two-component sensory transduction transcriptional |
| M. avium 104 | MAV_4209 | - | 8e-45 | 43.91% (230) | DNA-binding response regulator MtrA |
| M. smegmatis MC2 155 | MSMEG_0937 | regX3 | 5e-47 | 44.05% (227) | DNA-binding response regulator RegX3 |
| M. thermoresistible (build 8) | TH_4312 | - | 2e-45 | 42.73% (227) | PUTATIVE two component transcriptional regulator, winged helix |
| M. ulcerans Agy99 | MUL_4560 | regX3 | 9e-45 | 43.61% (227) | two component sensory transduction protein RegX3 |
| M. vanbaalenii PYR-1 | Mvan_1834 | - | 1e-113 | 79.84% (248) | two component transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_00773|M.leprae_Br4923 -------------MDIMRQRILVVDDDASLAEMLTIVLRGEGFDTAVIGD
MMAR_1299|M.marinum_M ----------------MRQRILVVDDDASLAEMLTIVLRGEGFDTAVIGD
Mb3274c|M.bovis_AF2122/97 -------------MDTMRQRILVVDDDASLAEMLTIVLRGEGFDTAVIGD
Rv3246c|M.tuberculosis_H37Rv -------------MDTMRQRILVVDDDASLAEMLTIVLRGEGFDTAVIGD
MAV_4209|M.avium_104 -------------MDSMRQRILVVDDDASLAEMLTIVLRGEGFDTAVIGD
MSMEG_0937|M.smegmatis_MC2_155 --------------------MLIVEDEESLADPLAFLLRKEGFEATVVGD
MUL_4560|M.ulcerans_Agy99 -----------------MTSVLIVEDEESLADPLAFLLRKEGFEATVVTD
TH_4312|M.thermoresistible__bu -----------------MTSVLIVEDEESLADPLAFLLRKEGFETTIVTD
Mflv_3887|M.gilvum_PYR-GCK MSTMD-HPQGSPSEHSRGYRALVVDDEVPLAEVVASYLQREQFDAVVADN
Mvan_1834|M.vanbaalenii_PYR-1 ---ME-REKAAPGGRAAGYRALVVDDEVPLAEVIAGYLEREQFETIVAAN
MAB_3339|M.abscessus_ATCC_1997 ---MDGSSRTAAAGPVPDYRALVVDDEVPLAEVVASYLKRESFEVHLAHD
*:*:*: .**: :: *. * *:. : :
MLBr_00773|M.leprae_Br4923 GTQALTAVRELRPDLVLLDLMLPGMNGIDVCRVLRADSGVPIVMLTAKTD
MMAR_1299|M.marinum_M GTQALTAVRELRPDLVLLDLMLPGMNGIDVCRVLRADSGVPIVMLTAKTD
Mb3274c|M.bovis_AF2122/97 GTQALTAVRELRPDLVLLDLMLPGMNGIDVCRVLRADSGVPIVMLTAKTD
Rv3246c|M.tuberculosis_H37Rv GTQALTAVRELRPDLVLLDLMLPGMNGIDVCRVLRADSGVPIVMLTAKTD
MAV_4209|M.avium_104 GTQALTAVRELRPDLVLLDLMLPGMNGIDVCRVLRADSGVPIVMLTAKTD
MSMEG_0937|M.smegmatis_MC2_155 GPSALAEFERSGADIVLLDLMLPGMSGTDVCKQLRARSSVPVIMVTARDS
MUL_4560|M.ulcerans_Agy99 GPAALAEFDRSGADIVLLDLMLPGMSGTDVCKQLRARSSVPVIMVTARDS
TH_4312|M.thermoresistible__bu GPSALAEFERSGADIILLDLMLPGMSGTDVCRRLRAKSKVPVIMVTARDS
Mflv_3887|M.gilvum_PYR-GCK GVDAIALARDFDPDVVILDLGLPGIDGLEVCRSLRTFSDAYVVMLTARDT
Mvan_1834|M.vanbaalenii_PYR-1 GLDAISVARELDPDVVILDLGLPGIDGLEVCRQIRTFSDAYVVMLTARDT
MAB_3339|M.abscessus_ATCC_1997 GRRALDLAREVDPDVVILDIGLPGVDGIEVCRQLRVFSDAYVVMLTARDT
* *: .*:::**: ***:.* :**: :*. * . ::*:**:
MLBr_00773|M.leprae_Br4923 TVDVVLGLESGADDYIMKPFKPKELVARVRARLRRNDD-------EPAEM
MMAR_1299|M.marinum_M TVDVVLGLESGADDYIMKPFKPKELVARVRARLRRNDD-------EPAEM
Mb3274c|M.bovis_AF2122/97 TVDVVLGLESGADDYIMKPFKPKELVARVRARLRRNDD-------EPAEM
Rv3246c|M.tuberculosis_H37Rv TVDVVLGLESGADDYIMKPFKPKELVARVRARLRRNDD-------EPAEM
MAV_4209|M.avium_104 TVDVVLGLESGADDYIMKPFKPKELVARVRARLRRNDD-------EPAEM
MSMEG_0937|M.smegmatis_MC2_155 EIDKVVGLELGADDYVTKPYSARELIARIRAVLRRGADN--DDAGADDGV
MUL_4560|M.ulcerans_Agy99 EIDKVVGLELGADDYVTKPYSARELIARIRAVLRRGGD---DDSEVSDGV
TH_4312|M.thermoresistible__bu EIDKVVGLELGADDYVTKPYSARELIARIRAVLRRGSDL--DGADVDDSV
Mflv_3887|M.gilvum_PYR-GCK ELDTVLGLTVGADDYVTKPFSPRELVARIRAMLRRPRLLQSPPTGADREV
Mvan_1834|M.vanbaalenii_PYR-1 EIDTVLGLTVGADDYVTKPFSPRELVARIRAMLRRPRVLHTAGAAAERNV
MAB_3339|M.abscessus_ATCC_1997 EVDTIVGLSVGADDYVTKPFSPREVVARVRAMLRRPRRAPESPAAVTPEL
:* ::** *****: **:..:*::**:** *** :
MLBr_00773|M.leprae_Br4923 LSI---ADVDIDVPAHKVTRNGEHISLTPLEFDLLVALARKPRQVFTRDV
MMAR_1299|M.marinum_M LSI---ADVDIDVPAHKVTRNGEQISLTPLEFDLLVALARKPRQVFTRDV
Mb3274c|M.bovis_AF2122/97 LSI---ADVEIDVPAHKVTRNGEQISLTPLEFDLLVALARKPRQVFTRDV
Rv3246c|M.tuberculosis_H37Rv LSI---ADVEIDVPAHKVTRNGEQISLTPLEFDLLVALARKPRQVFTRDV
MAV_4209|M.avium_104 LSI---ADVEIDVPAHKVTRNGEQISLTPLEFDLLVALARKPRQVFTRDV
MSMEG_0937|M.smegmatis_MC2_155 LEA---GPVRMDVERHVVSVNGEPITLPLKEFDLLEYLMRNSGRVLTRGQ
MUL_4560|M.ulcerans_Agy99 LES---GPVRMDVERHVVSVNGDAITLPLKEFDLLEYLMRNSGRVLTRGQ
TH_4312|M.thermoresistible__bu LEA---GPVRMDVDRHVVTVRGEPITLPLKEFELLEYLLRNSGRVLTRGQ
Mflv_3887|M.gilvum_PYR-GCK TPPRRFGALSIDVAAREVRIADDPILLTRTEFDILQALSGRPGIVLSRRQ
Mvan_1834|M.vanbaalenii_PYR-1 TPPRQFGPLSIDVAAREVRIDGEPILLTRTEFDILEALSGHPGIVLSRRQ
MAB_3339|M.abscessus_ATCC_1997 SPPRVFGALRINVDGREVHVGERPVTLTRTEFDVLAALSSRPGIVLSRKQ
. : ::* : * : *. **::* * .. *::*
MLBr_00773|M.leprae_Br4923 LLEQVWGYRHPADTRLVNVHVQRLRAKVEKDPENPTVVLTVRGVGYKAGP
MMAR_1299|M.marinum_M LLEQVWGYRHPADTRLVNVHVQRLRAKVEKDPENPTVVLTVRGVGYKAGP
Mb3274c|M.bovis_AF2122/97 LLEQVWGYRHPADTRLVNVHVQRLRAKVEKDPENPTVVLTVRGVGYKAGP
Rv3246c|M.tuberculosis_H37Rv LLEQVWGYRHPADTRLVNVHVQRLRAKVEKDPENPTVVLTVRGVGYKAGP
MAV_4209|M.avium_104 LLEQVWGYRHPADTRLVNVHVQRLRAKVEKDPENPTVVLTVRGVGYKAGP
MSMEG_0937|M.smegmatis_MC2_155 LIDRVWGADYVGDTKTLDVHVKRLRSKIEEDPANPVHLVTVRGLGYKLEG
MUL_4560|M.ulcerans_Agy99 LIDRVWGADYVGDTKTLDVHVKRLRSKIEADPANPVHLVTVRGLGYKLEG
TH_4312|M.thermoresistible__bu LIDRVWGADYVGDTKTLDVHVKRLRSKIEEDPANPVHLVTVRGLGYKLEG
Mflv_3887|M.gilvum_PYR-GCK LLEIIRDEPWVGNDQLVDVHIGHLRRKLGDDAARPRYIATVRGVGYRMGT
Mvan_1834|M.vanbaalenii_PYR-1 LIEIVRDGPWVGNEHLVDVHIGHLRRKLGDDAAQPRYIRTVRGVGYRMGT
MAB_3339|M.abscessus_ATCC_1997 LLDTVWGDTWVGNDNIVDVHVGHLRQKLGDSPSQPRYVMTVRGVGYRMGT
*:: : . .: . ::**: :** *: .. .* : ****:**:
MLBr_00773|M.leprae_Br4923 P----
MMAR_1299|M.marinum_M P----
Mb3274c|M.bovis_AF2122/97 P----
Rv3246c|M.tuberculosis_H37Rv P----
MAV_4209|M.avium_104 P----
MSMEG_0937|M.smegmatis_MC2_155 -----
MUL_4560|M.ulcerans_Agy99 -----
TH_4312|M.thermoresistible__bu -----
Mflv_3887|M.gilvum_PYR-GCK GA---
Mvan_1834|M.vanbaalenii_PYR-1 GSGAD
MAB_3339|M.abscessus_ATCC_1997 GS---