For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MKRSSWHLRAGSVVLGWLVALVMAAIAHPYIALSNWLLIHLLGLGAASNAILIWSRHFADALLRRSPDPP YPGQVIRLLAFNIGAVTVITGMLGGWWPVVLAGGVLVAAVAAVHAGDLMRFLRAALPARFSITVHYYVAA AACLAVGAGLGVAMANSALPGILAERFRTAHAVLNLFGWVGLTVLGTLVTLWPTMLRTRMADGVERAARR GLPALVLSVAVAVAGAILGSSRVVGVGALSFLTAVGFVLWPHIDEVRRKRPADFPTLSVLCGVVWLAGSL GYLSVGLLTAPNSMAAATVTAAAGPALLAGFLVQVLFGSLAYLIPMVVGGRASALAATAELERGAPWRLS VANFGLLICVLPVPNLVRVAVSALVLVAYGAFLPLLVRAVWLAQRNTGVRSTPGRLDPPPLRQRLGIAAA GFGVVILTAAAGVAADPATLGIGSAPTVTAAPTGHTTEVRVRVDGMRYVPDTITVPAGDRLRITFHNTGT DRHDLVLANGARTDRVAPGVTAVLDAGVIGSDLAGWCSIAGHRQMGMTLTIKAIGTPPVSGHEHELPGHG PDPSLTATDIARSLGGRPGPGFAARDPKLSAAETAEHRVTLTVSEVEREVSPGITQRLWTFGGSAPGPTL RGRVGDVFEITLINDGTIGHSIDFHAGSLAPDRPMRTIEPGAQLVYRFTATRAGVWLYHCATMPMSVHIA NGMFGAVVIDPPDLSTVDREYLMVQSEFYLGADGGEVDADKVAADTPDLVVFNGYAQQYDHAPLTARVGE RVRIWVLAAGPNRGSAFHVVGGQFDTVWAEGAYRLRPGAGGSQTLGLFPAQGGFVELSFPEPGNYPFVSH AMVDAERGAHGVIAVGN
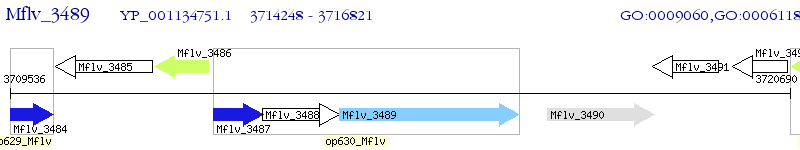
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3489 | - | - | 100% (857) | multicopper oxidase, type 3 |
| M. gilvum PYR-GCK | Mflv_2663 | - | 2e-06 | 25.65% (421) | ComEC/Rec2-related protein |
| M. gilvum PYR-GCK | Mflv_3485 | - | 2e-06 | 34.51% (113) | hypothetical protein Mflv_3485 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0980 | - | 4e-08 | 28.15% (302) | integral membrane protein |
| M. tuberculosis H37Rv | Rv0955 | - | 4e-08 | 28.15% (302) | integral membrane protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_0463c | - | 2e-09 | 24.54% (216) | oxidase (copper-binding protein) |
| M. marinum M | MMAR_4544 | - | 3e-08 | 28.86% (246) | integral membrane protein |
| M. avium 104 | MAV_1079 | - | 6e-07 | 29.28% (263) | hypothetical protein MAV_1079 |
| M. smegmatis MC2 155 | MSMEG_5517 | - | 5e-05 | 28.11% (249) | hypothetical protein MSMEG_5517 |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | MUL_4714 | - | 3e-08 | 28.86% (246) | integral membrane protein |
| M. vanbaalenii PYR-1 | Mvan_3626 | - | 0.0 | 44.69% (857) | multicopper oxidase, type 3 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3489|M.gilvum_PYR-GCK ------MKRSSWH----LRAGSVVLGWLVALVMAAIAHPYIALSNWLLIH
Mvan_3626|M.vanbaalenii_PYR-1 MTAQAPRRRNSLRGPVFLWANVVVLGWLAVSVGLLAAHSWLGLPGWVAVH
MMAR_4544|M.marinum_M --------------------------------------------------
MUL_4714|M.ulcerans_Agy99 --------------------------------------------------
MAV_1079|M.avium_104 --------------------------------------------------
Mb0980|M.bovis_AF2122/97 --------------------------------------------------
Rv0955|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_5517|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0463c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_3489|M.gilvum_PYR-GCK LLGLGAASNAILIWSRHFADALLRRSPDPPYPGQVIRLLAFNIGAVTVIT
Mvan_3626|M.vanbaalenii_PYR-1 ALLLGAVTNAIVIWSEHFAVAWCR-MPSPPQRRLALGLTALNLCVIAVLV
MMAR_4544|M.marinum_M -----------MTRVEDSRAAGARQARDLVKVAFAPAVVALVIIAAVTLI
MUL_4714|M.ulcerans_Agy99 --------------MEDSRAAGARQARDLVKVAFAPAVVALVIIAAVTLI
MAV_1079|M.avium_104 --------------MEDNRAAGARQARDLVRVAFAPAVVALVIIAAVTLI
Mb0980|M.bovis_AF2122/97 --------MNRVSASADDRAAGARPARDLVRVAFGPGVVALGIIAAVTLL
Rv0955|M.tuberculosis_H37Rv --------MNRVSASADDRAAGARPARDLVRVAFGPGVVALGIIAAVTLL
MSMEG_5517|M.smegmatis_MC2_155 ---------------MNNGPVGTRQARELLRVAFGPSVVALVIIAAVVLL
MAB_0463c|M.abscessus_ATCC_199 ------------------MAVSRSQWGRRGFLQLGAATAAAGVLAACGTR
. * : .
Mflv_3489|M.gilvum_PYR-GCK GMLGGWWPVVLAGGVLVAAVAAVHAGDLMRFLR---AALPARFSITVHYY
Mvan_3626|M.vanbaalenii_PYR-1 GVSTDTAALTGVGGTGITVIAVVHTVHLRRATRSTRSEAPGTFGYLIGFY
MMAR_4544|M.marinum_M QLVIANSDMTGALGAIASMWLGVHQVPIS---------------------
MUL_4714|M.ulcerans_Agy99 QLVIANSDMTGALGAIASMWLGVHQVPIS---------------------
MAV_1079|M.avium_104 QLLIANSDMTGALGAIASMWLGVHQVPVS---------------------
Mb0980|M.bovis_AF2122/97 QLLIANSDMTGAWGAIASMWLGVHLVPIS---------------------
Rv0955|M.tuberculosis_H37Rv QLLIANSDMTGAWGAIASMWLGVHLVPIS---------------------
MSMEG_5517|M.smegmatis_MC2_155 QLVIANSDMTGAFGAIASMWLGVHQVPVS---------------------
MAB_0463c|M.abscessus_ATCC_199 PVRGPSGQPVGPSSPRVGALEAVRRDGGGR--------------------
: . . *:
Mflv_3489|M.gilvum_PYR-GCK VAAAACLAVGAGLGVAMANSALPGILAERFRTAHAVLNLFGWVGLTVLGT
Mvan_3626|M.vanbaalenii_PYR-1 GAAGIMLASGAVLGALLALGS--GQWYVRLWSAHVHLTLLGWVGLTVLGT
MMAR_4544|M.marinum_M -------IGGRELG-----------------------------VMPLFPA
MUL_4714|M.ulcerans_Agy99 -------IGGRELG-----------------------------VMPLFPA
MAV_1079|M.avium_104 -------IGGRELG-----------------------------VLPLLPV
Mb0980|M.bovis_AF2122/97 -------IGGRALG-----------------------------VMPLLPV
Rv0955|M.tuberculosis_H37Rv -------IGGRALG-----------------------------VMPLLPV
MSMEG_5517|M.smegmatis_MC2_155 -------IGGRELG-----------------------------VMPLLPV
MAB_0463c|M.abscessus_ATCC_199 -------LVPVNLAARP-------------------------LQLDLAGT
*. : : .
Mflv_3489|M.gilvum_PYR-GCK LVTLWPTMLRTRMADGVERAARRGLPALVLSVAVAVAGAILGSSRVVGVG
Mvan_3626|M.vanbaalenii_PYR-1 MLTLWPTTLRARIGSDTHTAARRALPVLCAGLALTAAGMLAAVVWLTAAG
MMAR_4544|M.marinum_M LLMVWGTARTTALATSPYSSW-----------------------------
MUL_4714|M.ulcerans_Agy99 MLMVWGTARTTALATSPYSSW-----------------------------
MAV_1079|M.avium_104 LLMVWATARNTARATSPHSSW-----------------------------
Mb0980|M.bovis_AF2122/97 LLMVWATARSTARATSPQSSG-----------------------------
Rv0955|M.tuberculosis_H37Rv LLMVWATARSTARATSPQSSG-----------------------------
MSMEG_5517|M.smegmatis_MC2_155 LAMVWGTAKTTAAATAPNSSW-----------------------------
MAB_0463c|M.abscessus_ATCC_199 TVSTWGYGDQIPGPTIRARKG-----------------------------
*
Mflv_3489|M.gilvum_PYR-GCK ALSFLTAVGFVLWPHIDEVRRKRPADFPTLSVLCGVVWLAGSLGYLSVGL
Mvan_3626|M.vanbaalenii_PYR-1 LLCYAVGVGVVAVPLVRSAVARRPQGPAAWMLGAATGWLAAAVVYEAGRL
MMAR_4544|M.marinum_M --------------------------------------------------
MUL_4714|M.ulcerans_Agy99 --------------------------------------------------
MAV_1079|M.avium_104 --------------------------------------------------
Mb0980|M.bovis_AF2122/97 --------------------------------------------------
Rv0955|M.tuberculosis_H37Rv --------------------------------------------------
MSMEG_5517|M.smegmatis_MC2_155 --------------------------------------------------
MAB_0463c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_3489|M.gilvum_PYR-GCK LTAPNSMAAATVTAAAGPALLAGFLVQVLFGSLAYLIPMVVG-GRASALA
Mvan_3626|M.vanbaalenii_PYR-1 LVAGSVQALPAVTAETLPILIVGFTAQVLMGALTQLLPMLVGRGPAEHKV
MMAR_4544|M.marinum_M ----LVVRWVIASALGGPLLIAAICLAVIH--------------------
MUL_4714|M.ulcerans_Agy99 ----LVVRWVIASALGGPLLIAAICLAVIH--------------------
MAV_1079|M.avium_104 ----LVIRWVVASALGGPLLMTAVALAVIH--------------------
Mb0980|M.bovis_AF2122/97 ----LVVRWVVASALGGPLLMAAIALAVIH--------------------
Rv0955|M.tuberculosis_H37Rv ----LVVRWVVASALGGPLLMAAIALAVIH--------------------
MSMEG_5517|M.smegmatis_MC2_155 ----FVTRWVVASALGGPLLIAALLLAVIH--------------------
MAB_0463c|M.abscessus_ATCC_199 ----EVLRVAVANGLPAPTTVHWHGIALRN--------------------
... * : :
Mflv_3489|M.gilvum_PYR-GCK ATAELERGAPWRLSVANFGLLICVLPVPNLVRVAVSALVLVAYGAFLPLL
Mvan_3626|M.vanbaalenii_PYR-1 VAELLCAGWPARVAAINVAVPLVAGNWPDPLPLLGWVLAAAAVATFVVQA
MMAR_4544|M.marinum_M ----------------DASSVITELQTPSALRAFVSVLAVHAIGAGLGVW
MUL_4714|M.ulcerans_Agy99 ----------------DASSVITELQTPNALRAFVSVLAVHAIGAGLGVW
MAV_1079|M.avium_104 ----------------DASSVITELQTPSALRAFLSVLVVHAIGAGIGVW
Mb0980|M.bovis_AF2122/97 ----------------DASSVVTELQTPSALRAFTSVLVVHSVGAATGVW
Rv0955|M.tuberculosis_H37Rv ----------------DASSVVTELQTPSALRAFTSVLVVHSVGAATGVW
MSMEG_5517|M.smegmatis_MC2_155 ----------------DAASVITELQTPSALRAFGGVLAVHAIGALIGVS
MAB_0463c|M.abscessus_ATCC_199 ----------------------DMDGVPDLTQPQIAAAGSFTYEFALAHA
*. . :
Mflv_3489|M.gilvum_PYR-GCK VRAVWLAQRNTGVRSTPGRLDPPPLRQRLGIAAAGFGVVILTAAAGVAAD
Mvan_3626|M.vanbaalenii_PYR-1 LRVALPMARRGSIS-----------AQRQRIPGTGTGVVAGLAVVVLAVA
MMAR_4544|M.marinum_M SRVGRRALAVSPLP------------------------------------
MUL_4714|M.ulcerans_Agy99 SRVGIRALAVSPLP------------------------------------
MAV_1079|M.avium_104 SQVGPRALAASSLP------------------------------------
Mb0980|M.bovis_AF2122/97 SRVGRRALAATALP------------------------------------
Rv0955|M.tuberculosis_H37Rv SRVGRRALAATALP------------------------------------
MSMEG_5517|M.smegmatis_MC2_155 TKIGRRTLAASPLP------------------------------------
MAB_0463c|M.abscessus_ATCC_199 GTYWFHPHVGTQLD------------------------------------
:
Mflv_3489|M.gilvum_PYR-GCK PATLGIGSAPTVTAAPTGHTTEVRVRVDGMRYVPDTITVPAGDRLRITFH
Mvan_3626|M.vanbaalenii_PYR-1 VAATGH-DAPTQTVT-SGPTRTVDVTLQAMRMNPDVVEVAAGARLVLRVT
MMAR_4544|M.marinum_M ------------------------------NWLGDSLRAAAAGVLALLGL
MUL_4714|M.ulcerans_Agy99 ------------------------------NWLGDSLRAAAAGVLALLGL
MAV_1079|M.avium_104 ------------------------------YWLGDSLRAATAGVLALLGL
Mb0980|M.bovis_AF2122/97 ------------------------------DWLHDSMRAAAAGVLALLGL
Rv0955|M.tuberculosis_H37Rv ------------------------------DWLHDSMRAAAAGVLALLGL
MSMEG_5517|M.smegmatis_MC2_155 ------------------------------KWLPDAVRAASAGVLALFAL
MAB_0463c|M.abscessus_ATCC_199 ------------------------------RGLYAPLIVEDPDERADYDT
: .
Mflv_3489|M.gilvum_PYR-GCK NTGTDRHDLVLANGARTDRVAPGVTAVLDAGVIGSDLAGWCSIAGHRQMG
Mvan_3626|M.vanbaalenii_PYR-1 NVDAMPHDLRVETGEKTPVLGQGETATLDLGVIDGPRQAWCTVAGHRAAG
MMAR_4544|M.marinum_M SGLVTAGSLVVHWATMQELYG-----------ITDSAFGQFSLTVLSVLY
MUL_4714|M.ulcerans_Agy99 SGLVTAGSLVVHWATMQELYG-----------ITDSAFGQFSLTVLSVLY
MAV_1079|M.avium_104 SGFVTAGSLVVHWATMQELYG-----------ITDSIFGEFSLTVLSMLY
Mb0980|M.bovis_AF2122/97 SGVVTAGSLVVHWATMQELYG-----------ITDSIFGQFSLTVLSVLY
Rv0955|M.tuberculosis_H37Rv SGVVTAGSLVVHWATMQELYG-----------ITDSIFGQFSLTVLSVLY
MSMEG_5517|M.smegmatis_MC2_155 SGIVTAGSLVVHWGTMHDLFA-----------ITDTVFGQLSLTGLSVLY
MAB_0463c|M.abscessus_ATCC_199 ELVVALDDWIDGTGTDPDKVLAG---------LRDKGMPGMGHSMGDMPG
. . . . : . :
Mflv_3489|M.gilvum_PYR-GCK MTLTIKAIGTPPVSGHEHELPGHGPDPSLTATDIARSLGGRPGPGFAARD
Mvan_3626|M.vanbaalenii_PYR-1 MTMTIRTDGGETASTHDTHSPGDMG----AAMPAALDLGADPSPGWTPRD
MMAR_4544|M.marinum_M VPNVVVG-------------------------AAAIAVGASAHIGFATFS
MUL_4714|M.ulcerans_Agy99 VPNVVVG-------------------------AAAIAVGASAHIGFATFS
MAV_1079|M.avium_104 APNVIVG-------------------------TAAVAVGSSAHIGFATFS
Mb0980|M.bovis_AF2122/97 APNVIVG-------------------------TSAIAVGSSAHIGFATFS
Rv0955|M.tuberculosis_H37Rv APNVIVG-------------------------TSAIAVGSSAHIGFATFS
MSMEG_5517|M.smegmatis_MC2_155 IPNVIVG-------------------------TSAIAVGSSAHIGLATFS
MAB_0463c|M.abscessus_ATCC_199 MSIGSMG------------------------VSAQAPLGADGGDVTYPYY
. :*. .
Mflv_3489|M.gilvum_PYR-GCK PKLSAAETAEHRVTLTVSEVEREVSPGITQRLWTFGGSAPGPTLRGRVGD
Mvan_3626|M.vanbaalenii_PYR-1 ARLQPVAGTVHKIELTVAERDVEVAPGRFERRWTFGGSAPGPTLHGRVGD
MMAR_4544|M.marinum_M -------------AFTVFGGDIPAVP--------ILAAAPTPLLG----P
MUL_4714|M.ulcerans_Agy99 -------------AFTVFGGDIPAVP--------ILAAAPTPPLG----P
MAV_1079|M.avium_104 -------------SFTVFGGDIPALP--------VLAAVPRPPLG----P
Mb0980|M.bovis_AF2122/97 -------------SFAVLGGDIPALP--------ILAAAPTPPLG----P
Rv0955|M.tuberculosis_H37Rv -------------SFAVLGGDIPALP--------ILAAAPTPPLG----P
MSMEG_5517|M.smegmatis_MC2_155 -------------SFTVFGGDIPAVP--------VLAAAPTPPLG----P
MAB_0463c|M.abscessus_ATCC_199 LING---------RVAADPQSVDYRPG-TRVRLRIINAAGDSAFRVAIPG
.:. . * . :. . :
Mflv_3489|M.gilvum_PYR-GCK VFEITLINDGTIGHSIDFHAGSLAPDRPMRTIEPGAQLVYRFTATRAGVW
Mvan_3626|M.vanbaalenii_PYR-1 QFEITLVNDGTMGHSIDFHAGMLAPDVPMRTIAPGERLTYRFTAHHAGAW
MMAR_4544|M.marinum_M VWVALLIIGASSGVAVGQQC-------ARRPLPPLPAMGKLLAAALVGAL
MUL_4714|M.ulcerans_Agy99 VWVALLIIGASSGVAVGQQC-------ARRPLPPLPAMGKLLAAALLGAL
MAV_1079|M.avium_104 VWVALLIIGASSGVAVGQQC-------ARRPLPPVAAMVKLLAASVAGAL
Mb0980|M.bovis_AF2122/97 AWVALLIVGASSGVAVGQQC-------ARRALPFVAAMAKLLVAAVAGAL
Rv0955|M.tuberculosis_H37Rv AWVALLIVGASSGVAVGQQC-------ARRALPFVAAMAKLLVAAVAGAL
MSMEG_5517|M.smegmatis_MC2_155 VWVALLIVGAASAVALGQQC-------AARPLTPFAALAKLIVASLISAV
MAB_0463c|M.abscessus_ATCC_199 TPLTVTHTDGFPVQPRQTDALLIGMGERIDAIITVGEVSVPLLAAPYGKD
.. . .. .: : : * .
Mflv_3489|M.gilvum_PYR-GCK LYHCATMPMSVHIANGMFGAVVIDPPDLSTVDREYLMVQSEFYLGADGGE
Mvan_3626|M.vanbaalenii_PYR-1 LYHCSTMPMALHIANGMFGAVIIDPPGLAPVDREYVLVSSQLYLGEPGSQ
MMAR_4544|M.marinum_M AMALLGYGGSGKLGN--FGAVGVDEGALLVGVFFWFAVVGGATLVMAGG-
MUL_4714|M.ulcerans_Agy99 AMALLGYGGSGKLGN--FGAVGVDEGALLVGVFFWFAVVGGATLVMAGG-
MAV_1079|M.avium_104 VMSLLAYAGSGQLGN--FGHVGVDQGALLVGVFFWFAVVGGATVVMSGG-
Mb0980|M.bovis_AF2122/97 VMAVLGYGGGGRLGN--FGDVGVDEGALVLGVLFWFTFVGWVTVVIAGG-
Rv0955|M.tuberculosis_H37Rv VMAVLGYGGGGRLGN--FGDVGVDEGALVLGVLFWFTFVGWVTVVIAGG-
MSMEG_5517|M.smegmatis_MC2_155 TMALLGYAGSGRLGN--FGHVGVDQSTFGPAVFLWFVGIGGLTVVMSGG-
MAB_0463c|M.abscessus_ATCC_199 GYAQLVLRSSGTPVSGSAAEAAAALPKLAALDTASLSATEEVTLAVGEPD
. . . . : . :
Mflv_3489|M.gilvum_PYR-GCK VDADKVAADTPDLVVFNGYAQQYDHAPLTARVGERVRIWVLAAGPNRGSA
Mvan_3626|M.vanbaalenii_PYR-1 TQTDNLRAGQPDGWAFNGMAAQYDHAPLTAVVGERARIWVVNAGPGDSTA
MMAR_4544|M.marinum_M ---------------IRRRPRRPKPRPAPPVEADVDEPIH-EPVDEPFVP
MUL_4714|M.ulcerans_Agy99 ---------------IRRRPRRPKPRPVPPAEADVDEPIH-EPVDEPFVP
MAV_1079|M.avium_104 ---------------IRRPTRRLKPRPAP--AAPVAEEHD-SVFGEAVGP
Mb0980|M.bovis_AF2122/97 ---------------ISRRPKR--LRPAPPVELDADE-------SSPPVD
Rv0955|M.tuberculosis_H37Rv ---------------ISRRPKR--LRPAPPVELDADE-------SSPPVD
MSMEG_5517|M.smegmatis_MC2_155 ---------------IVRRVRVAPAAPPPAPEPEPEPVVPGEVLGEPAEA
MAB_0463c|M.abscessus_ATCC_199 ITYDLRLAGPGNKYSWTINEKTYNPAEGLPVREGQRVRLRFINNSMMFHP
Mflv_3489|M.gilvum_PYR-GCK FHVVGGQFDTVWAEGAYRLRP-----GAGGSQTLGLFPAQGGFVELSFP-
Mvan_3626|M.vanbaalenii_PYR-1 FHVVGGQFDTVYSEGAWLLRPSVGTEKSGAAQVLNLAPAQGGFVELTFP-
MMAR_4544|M.marinum_M LFDQDAPLGDMTSREPVAP----------GGDDADVPPAAPDEADWS---
MUL_4714|M.ulcerans_Agy99 LFDQDAPLDDVTSREPVAP----------GGDDADVPPAAPDEVDWS---
MAV_1079|M.avium_104 VGDVAG-LDEIAAEGVDEV----------AAEGVDESPGDAGEREGD---
Mb0980|M.bovis_AF2122/97 MFDGAA------SEQPPAS----------VAE--DVPPSHDDIANGL---
Rv0955|M.tuberculosis_H37Rv MFDGAA------SEQPPAS----------VAE--DVPPSHDDIANGL---
MSMEG_5517|M.smegmatis_MC2_155 DEPDDEPFGDEPYEETPEEPL--------VSWHADEPPEPAETDDDAGAP
MAB_0463c|M.abscessus_ATCC_199 MHLHGHTFAVRTASGAYGAR----------KDTVLVAPMQTVEVDFDAD-
* :
Mflv_3489|M.gilvum_PYR-GCK -----------EPGNYPFVSHAMVDAERGAHGVIAVGN
Mvan_3626|M.vanbaalenii_PYR-1 -----------EAGNYPFVDHDMRHGENGAHGVIAVAG
MMAR_4544|M.marinum_M -----------DGPDRPDGPDRQDGPDASHPPEHLT--
MUL_4714|M.ulcerans_Agy99 -----------DGPD------RQDGPDTSHPPEHLT--
MAV_1079|M.avium_104 -----------EGDP-----GERGSGDAGAAGERRPGE
Mb0980|M.bovis_AF2122/97 -----------KAPT----ADDEALPLSDEPPPRAD--
Rv0955|M.tuberculosis_H37Rv -----------KAPT----ADDEALPLSDEPPPRAD--
MSMEG_5517|M.smegmatis_MC2_155 VAPRSPRDLLDEDPEDHFFVDEFGDEEVSGDKPRNAED
MAB_0463c|M.abscessus_ATCC_199 -----------NPGQWLTHCHNIYHGEAGMMTVVSYRQ
. .