For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RHCSARRGRKRLPDATAGTLLIDARDIDDAEAQISAAFTTFRIRAAAPARSTRIQVWRNYAGSMGVDDVE YTYDLTYDMEAPEQILLCRMLSGVFEETHHGQPTRRHGIGTAIAFGAIQSPSSGRLEKTRYHMISVPREA LGQVAGDHPGSDAVRLTSTAPFSDVANQLVVDVADHIRHGLMSNPRAVVEPLVVGNATRYLAASMLAAFP HTGADRPPGETDNYDALRRGTAFIDDNAHRDISPADVAHAARVSGPTLKELFVRHRNCTPLQRLQRVRLE HVHRELAMADPQTSTVADIARKWGFWRLGPFTTVYRETYGQVPDRTLDQ*
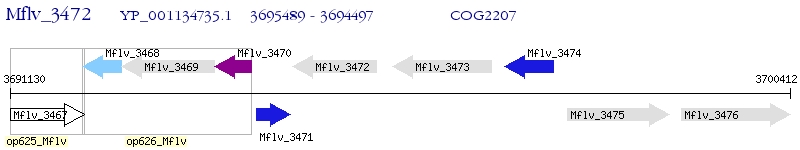
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3472 | - | - | 100% (330) | helix-turn-helix domain-containing protein |
| M. gilvum PYR-GCK | Mflv_0731 | - | 3e-39 | 33.02% (315) | helix-turn-helix domain-containing protein |
| M. gilvum PYR-GCK | Mflv_2365 | - | 1e-27 | 35.15% (202) | hypothetical protein Mflv_2365 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2899c | - | 5e-05 | 31.52% (92) | AraC family transcriptional regulator |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_4063 | - | 4e-23 | 28.36% (275) | transcriptional regulator, AraC family protein |
| M. smegmatis MC2 155 | MSMEG_2260 | - | 2e-10 | 28.26% (138) | putative transcriptional regulator |
| M. thermoresistible (build 8) | TH_3937 | - | 4e-29 | 37.99% (229) | PUTATIVE putative transcriptional regulator, AraC family |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0336 | - | 4e-62 | 41.48% (311) | helix-turn-helix domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3472|M.gilvum_PYR-GCK -----------------------MRHCSARRGRKRLPDATAGTLLIDARD
Mvan_0336|M.vanbaalenii_PYR-1 ---------------------------------------MSEAVIVDADN
TH_3937|M.thermoresistible__bu ----------------------VSDDTMVATAERQGPITRSRTELTDPSE
MAV_4063|M.avium_104 MGLTTMTVRDDGAPVGAPAEAARSDYGPVDPVLAVNGERRARPVAVEYKQ
MSMEG_2260|M.smegmatis_MC2_155 --------MSRLTRLGFIDIRRTSDPVRRKVRSRGVPPALADHEIFYTED
MAB_2899c|M.abscessus_ATCC_199 ------MRSARGPHSGEPGRSGEPHSGRLGRSGEPHSGRLGRSGEPHSGR
.
Mflv_3472|M.gilvum_PYR-GCK IDDAEAQISAAFTTFRIRAAAPARSTR----IQVWRNYAGSMGVDDVEYT
Mvan_0336|M.vanbaalenii_PYR-1 IGEAEAALSAAYTAMRLVSDRDVPSGR----TRIFRTRVGPLTVDDAEFD
TH_3937|M.thermoresistible__bu VGDFLEAVYGVR-VSRVRPPRSHDGPL----LTHVRADAGSFIVDRLHIA
MAV_4063|M.avium_104 FRGADAQSARRFFATAYDPGWRIAGVTNHCVVSHRRSDTGSMTVDEVAIQ
MSMEG_2260|M.smegmatis_MC2_155 VREAARLVGRTLSPLTLTVDAADAGGFAAT-MHGVRLRNVSLLYLDLHVA
MAB_2899c|M.abscessus_ATCC_199 LGRSGEPHSVVVLALPGTIGFDLATAVEVFRRVYLADGSRPYLVRVCGTE
. .
Mflv_3472|M.gilvum_PYR-GCK YDLTYDMEAPEQILLCRMLSGVFEETHHGQPTRRHGIGTAIAFGAIQSP-
Mvan_0336|M.vanbaalenii_PYR-1 FHVIADMDPSEDVLLCRVRGGMLGGHLPGRSPERHGAGTVLAFGGRRGER
TH_3937|M.thermoresistible__bu GELHTTADPIHKVVTLWPTSGRVVGRCAGLEG-RAGAGEIILSG-QPDLV
MAV_4063|M.avium_104 GRMTLEIPAGDTVVVIQPRAGALN--VAGGPLATPDFPLLVAHG----MA
MSMEG_2260|M.smegmatis_MC2_155 ATLDIPESGRYFAVHMPLNGSATVTHPAAEFDAGTTRSLVTSPG---DPV
MAB_2899c|M.abscessus_ATCC_199 PVVSAGPFGIATDLGLEAVSEAETIVVPARDNPTAPLPEGVLDALRGAYE
: : . . .
Mflv_3472|M.gilvum_PYR-GCK SSGRLEKTRYHMISVPREALGQVAG---DHPGSDAVRLTSTAPFSDVANQ
Mvan_0336|M.vanbaalenii_PYR-1 IYGDIDHARYDTIVLDRNLLAQVASGGLDAAG-EPVDLTSLAPVSEEANR
TH_3937|M.thermoresistible__bu RQTHCDDLAATVILLDPALVAHVASGSAGDDAAAAIRFASLRPAVGAT-R
MAV_4063|M.avium_104 CVLHCHGARFDVVTIAAEVLQKVAAEWHTALS-QQTQFLNWRPRSRAAVR
MSMEG_2260|M.smegmatis_MC2_155 RMSFAHDSPQMLVRIEEEAMNAHLTRLLGRNLVGPLKFEPEFDLTTEAAT
MAB_2899c|M.abscessus_ATCC_199 RGTRIASICAGAFTLAEAGILDGKRATTHWLAADLFRERYPAVHLDADVL
. : :
Mflv_3472|M.gilvum_PYR-GCK LVVDVADHIRHGLMSNPRAVVEPLVVGN---ATRYLAASMLAAFPHTGAD
Mvan_0336|M.vanbaalenii_PYR-1 HLADVMSHLRRDVASNAHAASQPLVTGP---LLRYLAGCVLAAFPNTAAA
TH_3937|M.thermoresistible__bu LWQDTVRYVRDHVLVD-DTRATPLVVGQ---AARLLAAVTASAFPNSAAE
MAV_4063|M.avium_104 AWHRALDYAMLTLASP-DTARQPLIAAG---MAGLLAGALLECYPSNLTE
MSMEG_2260|M.smegmatis_MC2_155 RWHAAVQLIHTEVYHAGSLVQRGCAIGP---VEELAMSSLLYLQPSTYYS
MAB_2899c|M.abscessus_ATCC_199 YVDEGQVLTSAGAAAGLDLCLHIVARDHGAAVAAEAARVAVAPLHRDGGQ
Mflv_3472|M.gilvum_PYR-GCK RP---PGETDNYDALRRGTAFIDDNAHRDISPADVAHAARVSGPTLKELF
Mvan_0336|M.vanbaalenii_PYR-1 PP---RVQDSSPRLLERALAFIDDNADKDISLGDIAAAARVTPQSVAAMF
TH_3937|M.thermoresistible__bu PAG-QTVNGQRPPLLRRALDYIDANAAADISLADIAAAVHVTPRALQYMF
MAV_4063|M.avium_104 QD--PVSDLALPETLKEAVSFIHRHAAEDIGINDVAAAVHLTPRAVQYLF
MSMEG_2260|M.smegmatis_MC2_155 DITRPTPATPRRAVIQQAVDYIEDHLAEKITMESLAEALHMSVRSIQQGF
MAB_2899c|M.abscessus_ATCC_199 AQYVIRNRRRAEVSLGEILAWIEHNAHRELSLEDLATEASTSVRSLNRRF
: . :*. : .: .:* : :: *
Mflv_3472|M.gilvum_PYR-GCK VRHRNCTPLQRLQRVRLEHVHRELAMADPQT-STVADIARKWGFWRLGPF
Mvan_0336|M.vanbaalenii_PYR-1 RRHLSCTPKEYLRKVRLDHAHRDLIANNPAT-ATVADIAARWGFGVVEAF
TH_3937|M.thermoresistible__bu RRHLQTTPLQYLRRIRLHRAHHDLLAQTPAS-DTVGAIAARWGFAHAGRF
MAV_4063|M.avium_104 RRQLDTTPTEYMRRVRLSRAHQELVAATTAS-STVTEIAQRWGFAHTGRF
MSMEG_2260|M.smegmatis_MC2_155 RTELGMSPTTFIRERRLERVREELTDAIPSDGVTVTAVAERWGFHHLGSF
MAB_2899c|M.abscessus_ATCC_199 RAETGQTPMQWLTGVRVRHAQQLLER----SADSVEWIGREVGFGSPANF
. :* : *: :.:. * :* :. . ** *
Mflv_3472|M.gilvum_PYR-GCK TTVYRETYGQVPDRTLDQ------
Mvan_0336|M.vanbaalenii_PYR-1 AVYYRGRYGVHPETTLAS------
TH_3937|M.thermoresistible__bu AMQYRQTYGRSPHQTLRG------
MAV_4063|M.avium_104 AVLYRQTYGQSPHTTLRQQTAV--
MSMEG_2260|M.smegmatis_MC2_155 AVEYRKRWGETPSETLRR------
MAB_2899c|M.abscessus_ATCC_199 REQFRRLTGVSPLAYRNTFRAQSA
:* * *