For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DVSGRYVVNTSDLDEASGILAATYSRMRISPCTHTNAHRWIRIWRTRVGPMTLDDAEITADIHYELDAPQ CVLLCRIRAGSLDLRIGDQVTSFGRGELIAVVADGASMEGVARQTRCDLVTFDRTLWGEVTAERGDRRFG SPLKRFSSPINPSAGQQVADVIDHIRRFVANNPLVAREPLVAGSAARYLAATMLTAFPSAGFTV*
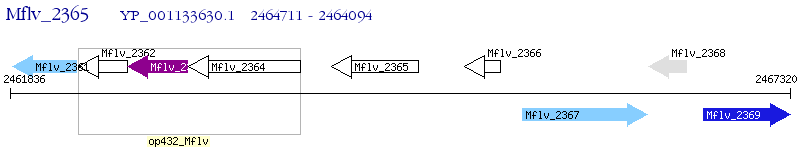
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2365 | - | - | 100% (205) | hypothetical protein Mflv_2365 |
| M. gilvum PYR-GCK | Mflv_3472 | - | 5e-28 | 35.15% (202) | helix-turn-helix domain-containing protein |
| M. gilvum PYR-GCK | Mflv_0731 | - | 5e-13 | 27.75% (191) | helix-turn-helix domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_4063 | - | 3e-05 | 26.45% (155) | transcriptional regulator, AraC family protein |
| M. smegmatis MC2 155 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_3937 | - | 5e-05 | 25.63% (199) | PUTATIVE putative transcriptional regulator, AraC family |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0336 | - | 1e-25 | 36.82% (201) | helix-turn-helix domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2365|M.gilvum_PYR-GCK ------------MDVSGRYVVNTSDLDEASGILAATY-SRMRISPCTH--
Mvan_0336|M.vanbaalenii_PYR-1 --------------MSEAVIVDADNIGEAEAALSAAY-TAMRLVSDRD--
TH_3937|M.thermoresistible__bu VSDDTMVATAERQGPITRSRTELTDPSEVGDFLEAVYGVRVSRVRPPR--
MAV_4063|M.avium_104 MGLTTMTVRDDGAPVGAPAEAARSDYGPVDPVLAVNGERRARPVAVEYKQ
. : . . * .
Mflv_2365|M.gilvum_PYR-GCK -------------------------TNAHRWIRIWRTRVGPMTLDDAEIT
Mvan_0336|M.vanbaalenii_PYR-1 -------------------------VPSGR-TRIFRTRVGPLTVDDAEFD
TH_3937|M.thermoresistible__bu -------------------------SHDGPLLTHVRADAGSFIVDRLHIA
MAV_4063|M.avium_104 FRGADAQSARRFFATAYDPGWRIAGVTNHCVVSHRRSDTGSMTVDEVAIQ
*: .*.: :* :
Mflv_2365|M.gilvum_PYR-GCK ADIHYELDAPQCVLLCRIRAGSLDLRIGDQVTS-FGRGELIAVVAD-GAS
Mvan_0336|M.vanbaalenii_PYR-1 FHVIADMDPSEDVLLCRVRGGMLGGHLPGRSPERHGAGTVLAFGGRRGER
TH_3937|M.thermoresistible__bu GELHTTADPIHKVVTLWPTSGRVVGRCAG-LEGRAGAGEIILSGQP-DLV
MAV_4063|M.avium_104 GRMTLEIPAGDTVVVIQPRAGALN------VAGGPLATPDFPLLVAHGMA
: . . *: .* : : .
Mflv_2365|M.gilvum_PYR-GCK MEGVARQTRCDLVTFDRTLWGEVTAERGDRRFGSPLKRFSSPINPSAGQQ
Mvan_0336|M.vanbaalenii_PYR-1 IYGDIDHARYDTIVLDRNLLAQVASGGLDAAGEPVDLTSLAPVSEEANRH
TH_3937|M.thermoresistible__bu RQTHCDDLAATVILLDPALVAHVASGSAGDDAAAAIRFASLRPAVGATRL
MAV_4063|M.avium_104 CVLHCHGARFDVVTIAAEVLQKVAAEWHTALSQQTQFLNWRPRSRAAVRA
: : : .*:: * :
Mflv_2365|M.gilvum_PYR-GCK VADVIDHIRRFVANNPLVAREPLVAGSAARYLAATMLTAFPSAGFTV---
Mvan_0336|M.vanbaalenii_PYR-1 LADVMSHLRRDVASNAHAASQPLVTGPLLRYLAGCVLAAFPNTAAAPP--
TH_3937|M.thermoresistible__bu WQDTVRYVRDHVLVDDTRA-TPLVVGQAARLLAAVTASAFPNSAAEPAGQ
MAV_4063|M.avium_104 WHRALDYAMLTLAS-PDTARQPLIAAGMAGLLAGALLECYPSNLTEQD-P
.: : : * **:.. **. .:*.
Mflv_2365|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0336|M.vanbaalenii_PYR-1 RVQDSSPRLLERALAFIDDNADKDISLGDIAAAARVTPQSVAAMFRRHLS
TH_3937|M.thermoresistible__bu TVNGQRPPLLRRALDYIDANAAADISLADIAAAVHVTPRALQYMFRRHLQ
MAV_4063|M.avium_104 VSDLALPETLKEAVSFIHRHAAEDIGINDVAAAVHLTPRAVQYLFRRQLD
Mflv_2365|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_0336|M.vanbaalenii_PYR-1 CTPKEYLRKVRLDHAHRDLIANNPATATVADIAARWGFGVVEAFAVYYRG
TH_3937|M.thermoresistible__bu TTPLQYLRRIRLHRAHHDLLAQTPASDTVGAIAARWGFAHAGRFAMQYRQ
MAV_4063|M.avium_104 TTPTEYMRRVRLSRAHQELVAATTASSTVTEIAQRWGFAHTGRFAVLYRQ
Mflv_2365|M.gilvum_PYR-GCK ----------------
Mvan_0336|M.vanbaalenii_PYR-1 RYGVHPETTLAS----
TH_3937|M.thermoresistible__bu TYGRSPHQTLRG----
MAV_4063|M.avium_104 TYGQSPHTTLRQQTAV