For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MRSKRLRAVAGAVFVATAASVQTVAGTPAAAADPCPDVEVIWARGTGAPAGLGWLGTAFVDSLRAKVGGR SVGAYGVNYPASFDFDASAPMGAADAAGRVQWMADNCPDTRLVLGGNSQGAGVIDLITLDATPRGRFTPT PLAPHLSDRVAAVAVFGNPLRDLPGGGPLSQVSGVFGSRSIDLCGLDDPFCSSGLNFPAHFSYIKNGMVE EATNFVAGRLQ
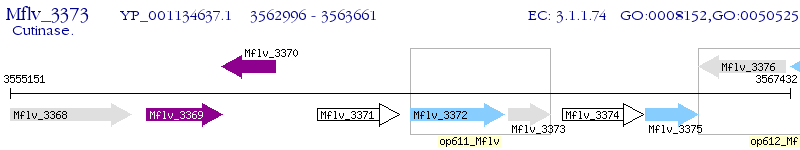
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3373 | - | - | 100% (221) | cutinase |
| M. gilvum PYR-GCK | Mflv_3552 | - | 6e-55 | 52.02% (198) | cutinase |
| M. gilvum PYR-GCK | Mflv_4965 | - | 1e-49 | 50.68% (219) | cutinase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2006c | cfp21 | 1e-50 | 46.88% (224) | cutinase precursor CFP21 |
| M. tuberculosis H37Rv | Rv1984c | cfp21 | 2e-50 | 46.88% (224) | cutinase precursor CFP21 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3766 | - | 8e-40 | 46.48% (213) | cutinase cut3 precursor |
| M. marinum M | MMAR_3837 | - | 3e-51 | 48.25% (228) | cutinase precursor |
| M. avium 104 | MAV_1682 | - | 2e-47 | 47.96% (221) | serine esterase, cutinase family protein |
| M. smegmatis MC2 155 | MSMEG_1528 | - | 3e-52 | 54.02% (224) | cutinase |
| M. thermoresistible (build 8) | TH_0238 | - | 2e-44 | 45.16% (217) | serine esterase, cutinase family protein |
| M. ulcerans Agy99 | MUL_1597 | cut1 | 7e-43 | 45.15% (206) | cutinase Cut1 |
| M. vanbaalenii PYR-1 | Mvan_3105 | - | 4e-90 | 69.09% (220) | cutinase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_3373|M.gilvum_PYR-GCK -----MRSKRLRAVAGAVFVAT------AASVQTVAGTPAAAADPCPDVE
Mvan_3105|M.vanbaalenii_PYR-1 -----MQVRRVLGIVGAAVAVS------AGLWPIGVGAPAASADPCPDIE
MAB_3766|M.abscessus_ATCC_1997 -MTTHQRGTRVGVVLSSFVVLTVLVLSAASVVGPVSAVPRASADPCSDVD
MSMEG_1528|M.smegmatis_MC2_155 -MSLDLVFRRARLVAAAALIATVMLAAPLAAHAPTGALGAARAADCPDIE
Mb2006c|M.bovis_AF2122/97 ------MTPRSLVRIVGVVVATT-----LALVSAPAGGRAAHADPCSDIA
Rv1984c|M.tuberculosis_H37Rv ------MTPRSLVRIVGVVVATT-----LALVSAPAGGRAAHADPCSDIA
MMAR_3837|M.marinum_M MCQHQNVNARQSTRLLGVGLATIC----AALLSAPLIIASASAAACPDAE
MAV_1682|M.avium_104 --------------MLGAGAMTAG----ATLLGAPLP---ASAAPCPDIE
TH_0238|M.thermoresistible__bu -LSRIGSAVGAALTTASLITGTVI----TSTVVAGTAVPAAAAQPCPDVQ
MUL_1597|M.ulcerans_Agy99 -------------------------------MFSAVAIPSASAEPCPDVE
* * *.*
Mflv_3373|M.gilvum_PYR-GCK VIWARGTGAPAGLGWLGTAFVDSLRAKVG--GRSVGAYGVNYPASFDFDA
Mvan_3105|M.vanbaalenii_PYR-1 VIFARGTGAEPGLGWVGDAFVNALRPKVG--GRSVGTYAVNYPAGFDFDN
MAB_3766|M.abscessus_ATCC_1997 VSFARGRKEPVGLGRVGDEFVDSLTPRVQATGATMNVYPVNYDAG--IFS
MSMEG_1528|M.smegmatis_MC2_155 VVFARGTDESPGIGRIGNAFVQALRGKVG--GRSVGTYAVNYPASYDFLA
Mb2006c|M.bovis_AF2122/97 VVFARGTHQASGLGDVGEAFVDSLTSQVG--GRSIGVYAVNYPASDDYRA
Rv1984c|M.tuberculosis_H37Rv VVFARGTHQASGLGDVGEAFVDSLTSQVG--GRSIGVYAVNYPASDDYRA
MMAR_3837|M.marinum_M VTFARGTAEPPGVGGVGQAFIDSLRSQLG--GRSLGVYAVNYPATEDWPP
MAV_1682|M.avium_104 VTFARGTAEPPGVGGVGQAFVEALRSQAG--ARSLGVYPVNYPADDDFAA
TH_0238|M.thermoresistible__bu VVFARGTGEPPGVGPTGQAFIDALRPRVG--DRSLDVYAVNYPAIDVWNT
MUL_1597|M.ulcerans_Agy99 VVFARGTGEPPGLGPTGQAFVNSLRSHIG--GRSLDVYPVNYPASDNWDT
* :*** *:* * *:::* : ::..* *** *
Mflv_3373|M.gilvum_PYR-GCK SAPMGAADAAGRVQWMADNCPDTRLVLGGNSQGAGVIDLITLDATPRGRF
Mvan_3105|M.vanbaalenii_PYR-1 SAPLGAADASGRVQWMAGNCPDTKLVLGGMSQGAGVIDLITVDPRPLGRF
MAB_3766|M.abscessus_ATCC_1997 AG-GGANDLSNHLQSVAASCPRTKLVIGGYSMGAEVIDTIIGVPNVGIGF
MSMEG_1528|M.smegmatis_MC2_155 AA-DGANDASGHVQWMVNNCPNTRLVLGGYSQGAAVIDVIAAVPFPAIGF
Mb2006c|M.bovis_AF2122/97 SASNGSDDASAHIQRTVASCPNTRIVLGGYSQGATVIDLSTSAMPPAV--
Rv1984c|M.tuberculosis_H37Rv SASNGSDDASAHIQRTVASCPNTRIVLGGYSQGATVIDLSTSAMPPAV--
MMAR_3837|M.marinum_M SASAGATDASTHVESMAASCPNTKLVLGGYSQGAMVIDLITIARAPVAGF
MAV_1682|M.avium_104 SASAGAGDASAHVQSMVADCPNTKLVLGGYSQGAMVIDLITIAQAPVAGL
TH_0238|M.thermoresistible__bu GID-GIRDAGAHVVRMAEQCPDTRLVLGGFSQGAAVMGFVTSAEVPAGVD
MUL_1597|M.ulcerans_Agy99 GLD-GIRDAGTHVVSMAGQCPQTKMVLGGYSQGTAVMSFVTSAAVPAGVD
. * * . :: . .** *::*:** * *: *:.
Mflv_3373|M.gilvum_PYR-GCK TP---TPLAPHLSDRVAAVAVFGNPLRDLP----GGGPLSQVSGVFGSRS
Mvan_3105|M.vanbaalenii_PYR-1 TP---TPMPPNVADHVAAVVVFGNPLRDIR----GGGPLPQMSGTYGSKS
MAB_3766|M.abscessus_ATCC_1997 N----RPLPPTVGDRIVAIATFGNATHRT------GGPLSGIAGVFGSRA
MSMEG_1528|M.smegmatis_MC2_155 N----APLPANVPEHVAAVAVFGNPSAKL------GLPLTASP-VYGSRA
Mb2006c|M.bovis_AF2122/97 ------------ADHVAAVALFGEPSSGFSSMLWGGGSLPTIGPLYSSKT
Rv1984c|M.tuberculosis_H37Rv ------------ADHVAAVALFGEPSSGFSSMLWGGGSLPTIGPLYSSKT
MMAR_3837|M.marinum_M ---IPQTLSAEVADHVAAVAVFGNPTD-----RYLGGPISEISPWYGHKA
MAV_1682|M.avium_104 ---IPQTLNADQAEHVSALALFGNPSD-----RYLGAPVSVVSPWYGAKA
TH_0238|M.thermoresistible__bu PATVPKPLDPDIAEHVAAVVLFGMPNAR--AMNFLGTPPVVIGPLYEDKT
MUL_1597|M.ulcerans_Agy99 PATVPKPLQPDVAEHVAAVVLFGMPNVR--AMNFLGEPAVEIGPAYQAKT
::: *:. ** . * . : ::
Mflv_3373|M.gilvum_PYR-GCK IDLCGLDDPFCSSG--LNFPA--------HFSYIK-NGMVEEATNFVAGR
Mvan_3105|M.vanbaalenii_PYR-1 IDLCAFDDPFCSPG--FNLPA--------HFAYVD-NGMVEEAANFAAGR
MAB_3766|M.abscessus_ATCC_1997 IDLCNQGDPICMAGPNNSWDA--------HTSYER-TGLPAEAAAFVASK
MSMEG_1528|M.smegmatis_MC2_155 IDLCNAGDPVCGSG--DSVPA--------HRAYD--GGPANQAAAFVAGL
Mb2006c|M.bovis_AF2122/97 INLCAPDDPICTGGGNIMA----------HVSYVQ-SGMTSQAATFAANR
Rv1984c|M.tuberculosis_H37Rv INLCAPDDPICTGGGNIMA----------HVSYVQ-SGMTSQAATFAANR
MMAR_3837|M.marinum_M IDLCAPNDPICTPG-ALALPSHDEMFSAAHLSYAQ-SGMPSQAATFVVSQ
MAV_1682|M.avium_104 IDLCAPGDPVCTPGGPLALPSHDEMVSPAHLSYRQ-SGMPAQAATFVAAH
TH_0238|M.thermoresistible__bu VKLCAVDDPVCSNGMNFAA----------HNTYATNAAMVDQGAAFAADR
MUL_1597|M.ulcerans_Agy99 IKVCVPEDPVCSDGLNFAA----------HNAYADDGAVVDQGVAFAASR
:.:* **.* * * :* . :.. *..
Mflv_3373|M.gilvum_PYR-GCK LQ------------------
Mvan_3105|M.vanbaalenii_PYR-1 LG------------------
MAB_3766|M.abscessus_ATCC_1997 LNRHGESEAE----------
MSMEG_1528|M.smegmatis_MC2_155 L-------------------
Mb2006c|M.bovis_AF2122/97 LDHAG---------------
Rv1984c|M.tuberculosis_H37Rv LDHAG---------------
MMAR_3837|M.marinum_M L-------------------
MAV_1682|M.avium_104 L-------------------
TH_0238|M.thermoresistible__bu LGVASDKPLPTPSSGQNFGN
MUL_1597|M.ulcerans_Agy99 LGGDPGGPVPAPSSG--FGN
*