For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTTHQRGTRVGVVLSSFVVLTVLVLSAASVVGPVSAVPRASADPCSDVDVSFARGRKEPVGLGRVGDEFV DSLTPRVQATGATMNVYPVNYDAGIFSAGGGANDLSNHLQSVAASCPRTKLVIGGYSMGAEVIDTIIGVP NVGIGFNRPLPPTVGDRIVAIATFGNATHRTGGPLSGIAGVFGSRAIDLCNQGDPICMAGPNNSWDAHTS YERTGLPAEAAAFVASKLNRHGESEAE
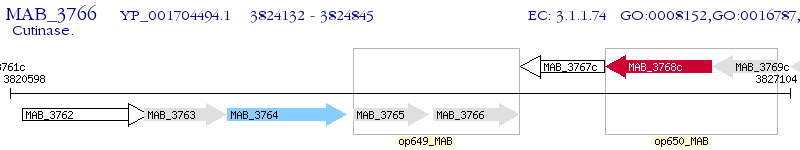
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3766 | - | - | 100% (237) | cutinase cut3 precursor |
| M. abscessus ATCC 19977 | MAB_3765 | - | 8e-57 | 54.39% (228) | cutinase cut3 precursor |
| M. abscessus ATCC 19977 | MAB_3763 | - | 7e-51 | 49.07% (214) | cutinase cut2 precursor |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3481 | cut3 | 3e-51 | 46.85% (222) | cutinase precursor CUT3 |
| M. gilvum PYR-GCK | Mflv_4965 | - | 2e-48 | 47.75% (222) | cutinase |
| M. tuberculosis H37Rv | Rv3451 | cut3 | 3e-51 | 46.85% (222) | cutinase precursor CUT3 |
| M. leprae Br4923 | - | - | - | - | - |
| M. marinum M | MMAR_1098 | cut3 | 8e-54 | 48.62% (218) | cutinase precursor, Cut3 |
| M. avium 104 | MAV_4394 | - | 1e-52 | 45.76% (236) | serine esterase, cutinase family protein |
| M. smegmatis MC2 155 | MSMEG_1528 | - | 4e-45 | 44.84% (223) | cutinase |
| M. thermoresistible (build 8) | TH_3765 | - | 2e-40 | 41.30% (230) | PUTATIVE Cutinase |
| M. ulcerans Agy99 | MUL_0856 | cut3 | 8e-53 | 48.17% (218) | cutinase precursor, Cut3 |
| M. vanbaalenii PYR-1 | Mvan_0371 | - | 1e-48 | 46.23% (212) | cutinase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_4965|M.gilvum_PYR-GCK -----MKIDLEVVTRGVR---ALVAAAVAAGVLVVAP-AVAAPR--SLPA
MSMEG_1528|M.smegmatis_MC2_155 -------MSLDLVFRRAR---LVAAAALIATVMLAAPLAAHAPTG-ALGA
Mvan_0371|M.vanbaalenii_PYR-1 -------MSERRSTGPAASDTSVRRALVMAGVGGVLAAGLIGPG--AASA
TH_3765|M.thermoresistible__bu MVGTMGEVNSDLELPGTRRTVALLTAALFLAAGLVTGSAVSPASAPLAPV
Mb3481|M.bovis_AF2122/97 -------MNNRPIRLLTSGRAGLGAGALITAVVLLIALGAVW---T--PV
Rv3451|M.tuberculosis_H37Rv -------MNNRPIRLLTSGRAGLGAGALITAVVLLIALGAVW---T--PV
MMAR_1098|M.marinum_M -------MNKHPLAVL--------AGALVTATGLLAAPSITG---TAIPS
MUL_0856|M.ulcerans_Agy99 -------MNKHPLAVL--------AGALVTATGLLAAPSITG---TAIPS
MAV_4394|M.avium_104 -------MKTHRIGRLLG----LGSSALITAAGLLGAP-------AAVPV
MAB_3766|M.abscessus_ATCC_1997 -------MTTHQRGTRVG---VVLSSFVVLTVLVLSAASVVGPV-SAVPR
: . : .
Mflv_4965|M.gilvum_PYR-GCK AGAQGCTDIQVVFARGTDEPPGLGRVGSAFVESL--RGRVGGRSVGSYAV
MSMEG_1528|M.smegmatis_MC2_155 ARAADCPDIEVVFARGTDESPGIGRIGNAFVQAL--RGKVGGRSVGTYAV
Mvan_0371|M.vanbaalenii_PYR-1 QAAESCPDVQVVFARGTAEPAGLGRVGQAFVDDL--QNELSGRTVSVYAV
TH_3765|M.thermoresistible__bu AQAAPCPDVEVIFARGRVESPGPGIIGNAFVNSL--RSKT-DKNVSLYAV
Mb3481|M.bovis_AF2122/97 AFADGCPDAEVTFARGTGEPPGIGRVGQAFVDSL--RQQT-GMEIGVYPV
Rv3451|M.tuberculosis_H37Rv AFADGCPDAEVTFARGTGEPPGIGRVGQAFVDSL--RQQT-GMEIGVYPV
MMAR_1098|M.marinum_M ASAADCPDAEVVFARGTSEPPGIGRVGEAFVDSL--RQQT-GKNIGVYAV
MUL_0856|M.ulcerans_Agy99 ASAADCPDAEVVFPRGTSEPPGIGRVGEAFVDSL--RQQT-GKNIGVYAV
MAV_4394|M.avium_104 ASADDCPDVEVIFARGTNEPPGMGRVGDAFVESL--RQQTSGLNIDTYGV
MAB_3766|M.abscessus_ATCC_1997 ASADPCSDVDVSFARGRKEPVGLGRVGDEFVDSLTPRVQATGATMNVYPV
* *.* :* *.** *. * * :*. **: * : . . :. * *
Mflv_4965|M.gilvum_PYR-GCK AYPATFDFLAAAG-GANDASNHIQWMMANCPSTRLVLGGYSQGAAVIDVI
MSMEG_1528|M.smegmatis_MC2_155 NYPASYDFLAAAD-GANDASGHVQWMVNNCPNTRLVLGGYSQGAAVIDVI
Mvan_0371|M.vanbaalenii_PYR-1 NYPASYDFLQSAPVGADDASARIQATVATCPATSIVIGGYSQGAAAVDLI
TH_3765|M.thermoresistible__bu QYPADY----EIDIGANDMSRRIQHMAATCPDTRLVLGGYSLGAAVTDVV
Mb3481|M.bovis_AF2122/97 NYAASRLQLHGGD-GANDAISHIKSMASSCPNTKLVLGGYSQGATVIDIV
Rv3451|M.tuberculosis_H37Rv NYAASRLQLHGGD-GANDAISHIKSMASSCPNTKLVLGGYSQGATVIDIV
MMAR_1098|M.marinum_M NYPASRLQLHGGD-GANDVISHVKSMASSCPNTKIVLGGYSQGADVIDIV
MUL_0856|M.ulcerans_Agy99 NYPASRLQLHGGD-GANDVISHVKSMASSCPNTKIVQGGYSQGADVIDIV
MAV_4394|M.avium_104 NYAASKLQLHGGD-GANDTINRIKQAVQKCPNTKIVLGGYSQGASVMDIV
MAB_3766|M.abscessus_ATCC_1997 NYDAG--IFSAGG-GANDLSNHLQSVAASCPRTKLVIGGYSMGAEVIDTI
* * **:* ::: .** * :* **** ** . * :
Mflv_4965|M.gilvum_PYR-GCK AAVPFPAVGFDRPLPPNAPDFIAAIALFGNPTTKVGLPITAS-PVWGTRS
MSMEG_1528|M.smegmatis_MC2_155 AAVPFPAIGFNAPLPANVPEHVAAVAVFGNPSAKLGLPLTAS-PVYGSRA
Mvan_0371|M.vanbaalenii_PYR-1 TADPNATFGFGRPLPPAVADHVAAVAVFGNPSTKIGRPLTAVSPLYGANA
TH_3765|M.thermoresistible__bu LAVPISALGFDSPLPAGVDRHIAAIALFGNGIAWAG-PISNFSPLYAERT
Mb3481|M.bovis_AF2122/97 AGVPLGSISFGSPLPAAYADNVAAVAVFGNPSNRAGGSLSSLSPLFGSKA
Rv3451|M.tuberculosis_H37Rv AGVPLGSISFGSPLPAAYADNVAAVAVFGNPSNRAGGSLSSLSPLFGSKA
MMAR_1098|M.marinum_M AGVPLAGISFGNELPAQYADNIAAVAVFGNVANRSGGSLTTQSSLLGSKS
MUL_0856|M.ulcerans_Agy99 AGVPLAGISFGNELPAQYADNIAAVAVFGNVANRSGGSLTTQSSLLGSKS
MAV_4394|M.avium_104 AGVPIGGINWGSSITPDLASHVAAVATFGDVADRAGGTLPSQSALLGSKA
MAB_3766|M.abscessus_ATCC_1997 IGVPNVGIGFNRPLPPTVGDRIVAIATFGNATHRTGGPLSGIAGVFGSRA
. * ..:. :.. :.*:* **: * .:. : . .:
Mflv_4965|M.gilvum_PYR-GCK IDLCNGADPVCSAGD--------DIAAHSNYGPAGFTDQAAAFVAGLV--
MSMEG_1528|M.smegmatis_MC2_155 IDLCNAGDPVCGSGD--------SVPAHRAYD-GGPANQAAAFVAGLL--
Mvan_0371|M.vanbaalenii_PYR-1 IDLCNGADPVCSSGD--------DRPAHSQYVQAGLAQQAADFVAGRVAG
TH_3765|M.thermoresistible__bu IELCHGADPICNPADPETWRDNWSDHAATKYVQSGMVDQAADFVAARL--
Mb3481|M.bovis_AF2122/97 IDLCNPTDPICHVGPGN------EFSGHIDGYIPTYTTQAASFVVQRLRA
Rv3451|M.tuberculosis_H37Rv IDLCNPTDPICHVGPGN------EFSGHIDGYIPTYTTQAASFVVQRLRA
MMAR_1098|M.marinum_M IDLCNPSDPICHSGPGN------DWNGHTDGYVPTYTTQAASFVAAKLAA
MUL_0856|M.ulcerans_Agy99 IDLCNPSDPICHSGPGN------DWNGHTDGYVPTYTTQAASFVAAKLAA
MAV_4394|M.avium_104 IDLCNPNDPICHAGPGN------EWSGHTEGYVPVYTTQAAAFVASKLMG
MAB_3766|M.abscessus_ATCC_1997 IDLCNQGDPICMAGPNN------SWDAHTSYERTGLPAEAAAFVASKLNR
*:**: **:* . . . :** **. :
Mflv_4965|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_1528|M.smegmatis_MC2_155 --------------------------------------------------
Mvan_0371|M.vanbaalenii_PYR-1 H-----------------SPSDVVINASAR--------------------
TH_3765|M.thermoresistible__bu --------------------------------------------------
Mb3481|M.bovis_AF2122/97 G--SVP----HLPGSVPQLPGSVLQMPG--TAAP-----APESLHGR---
Rv3451|M.tuberculosis_H37Rv G--SVP----HLPGSVPQLPGSVLQMPG--TAAP-----APESLHGR---
MMAR_1098|M.marinum_M S--SMAPV-VQLPGPVPRAPGPAPQAPGPATASPGTPVGATGTLVGAH--
MUL_0856|M.ulcerans_Agy99 S--SMAPV-VRLPGPVPRAPGPAPQAPGPATASPGTPVGATGTLVGAH--
MAV_4394|M.avium_104 AGQSVPGYGPSVPGAGPQSPGYGQQSPVYGQTPPGSGPSIHDNAPGPGTA
MAB_3766|M.abscessus_ATCC_1997 H-------------------GESEAE------------------------
Mflv_4965|M.gilvum_PYR-GCK ----------------
MSMEG_1528|M.smegmatis_MC2_155 ----------------
Mvan_0371|M.vanbaalenii_PYR-1 ----------------
TH_3765|M.thermoresistible__bu ----------------
Mb3481|M.bovis_AF2122/97 ----------------
Rv3451|M.tuberculosis_H37Rv ----------------
MMAR_1098|M.marinum_M ----------------
MUL_0856|M.ulcerans_Agy99 ----------------
MAV_4394|M.avium_104 PAPAVPSTPPSQAPLV
MAB_3766|M.abscessus_ATCC_1997 ----------------