For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VLWDMDGTLVDSEKLWDVSLSKLYETYDAEMSRETRTALVGASAEETMVTVYTELGLDLDPAAMAESIRW LHAHTAELFDEGLPWCDGAFELLERLVAEGTPMALVTNTNRHLADRALESIGRHYFSFTVCGDEVPRGKP APDPYLRAAELLALDPSDCLAVEDSVTGTAAAERAGCAVLVVPNDVPVPGGLRRRHVQTLADLGPADLRR IHAEITRELGERTA
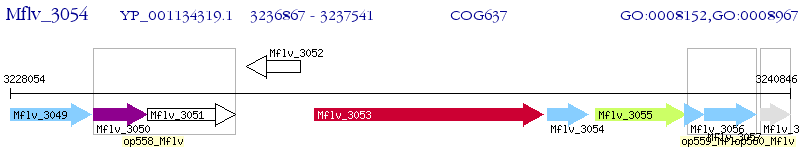
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_3054 | - | - | 100% (224) | HAD family hydrolase |
| M. gilvum PYR-GCK | Mflv_1072 | - | 2e-10 | 29.88% (241) | beta-phosphoglucomutase family hydrolase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3433 | - | 2e-13 | 28.02% (207) | hydrolase |
| M. tuberculosis H37Rv | Rv3400 | - | 2e-13 | 28.02% (207) | hydrolase |
| M. leprae Br4923 | MLBr_00393 | - | 7e-10 | 26.02% (196) | putative hydrolase |
| M. abscessus ATCC 19977 | MAB_2130 | - | 4e-74 | 61.86% (215) | putative hydrolase |
| M. marinum M | MMAR_0580 | - | 6e-12 | 28.35% (194) | hydrolase |
| M. avium 104 | MAV_4350 | - | 4e-10 | 25.63% (238) | beta-phosphoglucomutase hydrolase |
| M. smegmatis MC2 155 | MSMEG_4183 | - | 1e-78 | 66.06% (218) | phosphoglycolate phosphatase, chromosomal |
| M. thermoresistible (build 8) | TH_0959 | - | 9e-67 | 58.33% (216) | phosphoglycolate phosphatase, chromosomal |
| M. ulcerans Agy99 | MUL_0545 | - | 1e-11 | 27.84% (194) | hydrolase |
| M. vanbaalenii PYR-1 | Mvan_3473 | - | 1e-103 | 83.04% (224) | HAD family hydrolase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3433|M.bovis_AF2122/97 MANWYRPN-YPEVRSRVLGLPEKVRACLFDLDGVLTDTASLHTKAWKAMF
Rv3400|M.tuberculosis_H37Rv MANWYRPN-YPEVRSRVLGLPEKVRACLFDLDGVLTDTASLHTKAWKAMF
MAV_4350|M.avium_104 MVEWIPPDPNHRKRVPVLGLPEKVHACLFDLDGVLTDTASVHTKAWKAMF
MLBr_00393|M.leprae_Br4923 MANWDHPT--SKVRSTVLGLPEQVRACLFDLDGVLTDTASVHAKAWQTMF
Mflv_3054|M.gilvum_PYR-GCK --------------------------MLWDMDGTLVDSE--------KLW
Mvan_3473|M.vanbaalenii_PYR-1 -------MLRGSRRDTAACQNGRVRAVLWDMDGTLVDSE--------KLW
MSMEG_4183|M.smegmatis_MC2_155 -----------------------MLAVLWDMDGTLVDSE--------KLW
TH_0959|M.thermoresistible__bu ------------------------------MDGTLVDSE--------RLW
MAB_2130|M.abscessus_ATCC_1997 -----------------------MRAVLWDMDGTLIESE--------KLW
MMAR_0580|M.marinum_M ------------------MPELHAAALLFDMDGTLVDST--------AVV
MUL_0545|M.ulcerans_Agy99 ------------------MPELHAAALLFDVDGTLVDST--------AVV
:**.* :: :
Mb3433|M.bovis_AF2122/97 DAYLAERAERTGEKFVPFDPAADYHTYVDGKKREDGVRSFLSSRAIEIPD
Rv3400|M.tuberculosis_H37Rv DAYLAERAERTGEKFVPFDPAADYHTYVDGKKREDGVRSFLSSRAIEIPD
MAV_4350|M.avium_104 DAYLADRAKRTGETFVPFDPAADYRKYVDGKKREDGVRSFLQSRGIHLPD
MLBr_00393|M.leprae_Br4923 DTYLYQRAKHTGENFVPFDPTADYRQYVDGKKREDGVRSFLGSRGITLPE
Mflv_3054|M.gilvum_PYR-GCK DVSLSKLYETYDAEMSR-----ETRTALVGASAEETMVTVYTELGLDL--
Mvan_3473|M.vanbaalenii_PYR-1 DVSLSALYETFGGVMSR-----ETRATLVGASAEETMVTVYTELGLDL--
MSMEG_4183|M.smegmatis_MC2_155 DISMHALYARMGAVLTP-----EVRESTVGGSSETVMRIVYEDIGLEP--
TH_0959|M.thermoresistible__bu DLSLRELYRRLGRELRP-----EVRAATVGGAADTVMRIVYDDLGLDP--
MAB_2130|M.abscessus_ATCC_1997 DISMGELCRKLGGEMTP-----ELRTALVGGAADATMRMIFTALELEP--
MMAR_0580|M.marinum_M EATWGAFAELHSLDLAE------VLAFAHGRPTAATVRHFLS--------
MUL_0545|M.ulcerans_Agy99 EATWGAFAELHSLDLAE------VLAFAHGRPTAATVRHFLS--------
: . : * : .
Mb3433|M.bovis_AF2122/97 GSPDDPGAAETVYGLGNRKNDMLHKLLRDDGAQVFDGSRRYLEAVTAAGL
Rv3400|M.tuberculosis_H37Rv GSPDDPGAAETVYGLGNRKNDMLHKLLRDDGAQVFDGSRRYLEAVTAAGL
MAV_4350|M.avium_104 GSPDDAGDAETIYGLGNRKNDMFQKVLKQDGVKVFDGSRRYLEAASAAGL
MLBr_00393|M.leprae_Br4923 GSADNSGDVETIYGLGNRKDDLFRQVLKEHGIEVFDGSRRYLEAITYAGL
Mflv_3054|M.gilvum_PYR-GCK ---DPAAMAESIRWLHAHTAELF-----DEGLPWCDGAFELLERLVAEGT
Mvan_3473|M.vanbaalenii_PYR-1 ---DPEAMAESVRWLHDHTAELF-----DGGLPWCDGAREMLEGLAAERT
MSMEG_4183|M.smegmatis_MC2_155 ---DPAAMAESADWLHAYTGELF-----EQGLPWRPGAQEMLDALVAAEV
TH_0959|M.thermoresistible__bu ---DPEAMARTADWLHDYTGELF-----ASGLPWCAGARELLDELAAAAV
MAB_2130|M.abscessus_ATCC_1997 ---DPVRMASENDWLHQYTGELF-----ATGLIWCDGAQEMLGQLAAEQM
MMAR_0580|M.marinum_M ---HPALVASETRRLVEHEESTT------EGVREVPGAAEFIAALPRSAW
MUL_0545|M.ulcerans_Agy99 ---HPALVASETRRLVEHEESTT------EGVREVPGAAEFIAALPRSAW
. . * . * *: . :
Mb3433|M.bovis_AF2122/97 GVAVVSSSANTRDVLATTGLDRFVQQRVDGVTLREEHIAGKPAPDSFLRA
Rv3400|M.tuberculosis_H37Rv GVAVVSSSANTRDVLATTGLDRFVQQRVDGVTLREEHIAGKPAPDSFLRA
MAV_4350|M.avium_104 GIAVVSSSANTRDVLEVTGLDRFVQQRVDGITLREEHLAGKPAPDSFLRG
MLBr_00393|M.leprae_Br4923 GVAVVSSSTNTRDVLKITGLDRFVQQQVDGITLREEHIAGKPAPDSYLRG
Mflv_3054|M.gilvum_PYR-GCK PMALVT---NTNRHLADRALESIGRHYFSFTVCGDEVPRGKPAPDPYLRA
Mvan_3473|M.vanbaalenii_PYR-1 PAALVT---NTQRGLTERALNSIGRHYFSVTVCADEVASGKPAPDPYLRA
MSMEG_4183|M.smegmatis_MC2_155 PMALVT---NTRRDLAERALNSIGRHYFSVSVCGDEVPSGKPAPDPYLRA
TH_0959|M.thermoresistible__bu PMALVT---NTRRTLTEQALHSIGRHYFAVTVCGDEVPNGKPAPDIYHRA
MAB_2130|M.abscessus_ATCC_1997 PMALVT---NTIRSLTDQALKTIGTHWFTESVCGDEVPAGKPAPDPYLRA
MMAR_0580|M.marinum_M AVVT-----SASRVLASNRMRAAGIPLPDVLISADDITRGKPDPQGYLLA
MUL_0545|M.ulcerans_Agy99 AVVT-----SASRVLASNRMRAAGIPLPDVLISADDITRGKPDPQGYLLA
. .: * : :: *** *: : .
Mb3433|M.bovis_AF2122/97 AELLGVTPDAAAVFEDALSGVAAGRAGNFAVVVG-----INRTGRAAQAA
Rv3400|M.tuberculosis_H37Rv AELLGVTPDAAAVFEDALSGVAAGRAGNFAVVVG-----INRTGRAAQAA
MAV_4350|M.avium_104 AQLLDVPAGAAAVFEDALSGVAAGRAGNFGFVVG-----VDRVG---QAE
MLBr_00393|M.leprae_Br4923 AQLLDVAPDAAAVFEDALSGVQAGLSGHFGFVVG-----INRTGRVDQAE
Mflv_3054|M.gilvum_PYR-GCK AELLALDPSDCLAVEDSVTGTAAAERAGCAVLVVPNDVPVPGGLRRRHVQ
Mvan_3473|M.vanbaalenii_PYR-1 AELLGVDPADCLAVEDSVTGTAAAEKAGCAVLVVPNDVPVPGGLRRRHVG
MSMEG_4183|M.smegmatis_MC2_155 AELLGVPVARCLAVEDSVTGTTAAEAAGCPVLVIPNDVAVPHGPRRKQVE
TH_0959|M.thermoresistible__bu ADLLGVAPEACLAVEDSPTGAAAAEGAGCPVLVVPNALEVQHSPRRRQIP
MAB_2130|M.abscessus_ATCC_1997 AALLGVEPRDCLAIEDSVTGAAAAEAAGCAVLVVPNHVEVPAGARRRHAA
MMAR_0580|M.marinum_M ADALRVNIGSTIVFEDSPAGIRAGIASNALTVAIG--TLAAFDGHIERFE
MUL_0545|M.ulcerans_Agy99 ADALRVNIGSTIVFEDSPAGIRAGIASNALTVAIG--TLAAFDGHIERFE
* * : ..**: :* *. . :. :
Mb3433|M.bovis_AF2122/97 QLRRHGADVVVTDLAELL--------------
Rv3400|M.tuberculosis_H37Rv QLRRHGADVVVTDLAELL--------------
MAV_4350|M.avium_104 DLRRNGADVVVSDLAELLAS------------
MLBr_00393|M.leprae_Br4923 ELRRIGANVVVTDLAELL--------------
Mflv_3054|M.gilvum_PYR-GCK TLADLGPADLRRIHAEITRELGERTA------
Mvan_3473|M.vanbaalenii_PYR-1 SLAGLDVADLRRIYAEIDRELRERTA------
MSMEG_4183|M.smegmatis_MC2_155 SLSLLGVDDLRAIHAELGPE---RLSA-----
TH_0959|M.thermoresistible__bu SLTHVDVPLMREIHADLSAAPGPGVPAGHRPL
MAB_2130|M.abscessus_ATCC_1997 SLSGLSVRDLRDAHADVLAATTCKPI------
MMAR_0580|M.marinum_M NFHGFEVAGQPPAGGVILSMPAPMNCGV----
MUL_0545|M.ulcerans_Agy99 NFHGFEVAGQPPAGGVILSMPAPMNCGV----
: . :