For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
NAMPDILITACPPVGHIGPLLNVAHGLVAHGHRVTVLTSARHADKIRAVGADPRPLPYGADYDDARLDAD LPGRAHTSGIARINFDVEHVFVRPMPYQFSAVQELLAANSFDAVITDAFFLGVIPLLLDPTLARPPIMAY STTPLFLSSRDTAPAGPGILPMPGAVGRLRNRLLGLLSRAVLLRPAHRSAERMLRAMGLPDLPVFVLECA ILADRVIVPTVPDFEYPRRDLPAHVRFVGAVSPLPGDDFIAPAWWGELYRGRPVVHVTQGTVDNADLTRL IEPTIEALAGEDVTVVVSTGGRPVSQIRTPLPANTFVAEFLPHDVLLPLVDVLVTNGGYGAVQRALAEGV PLVVAGDTEDKPEVAARVEYFGVGVNLRTGTPAPQDLRDAVNAVLSDAGYRTRAGRLQAAYASRDSVTAI AALLDEVIPEQAAAR*
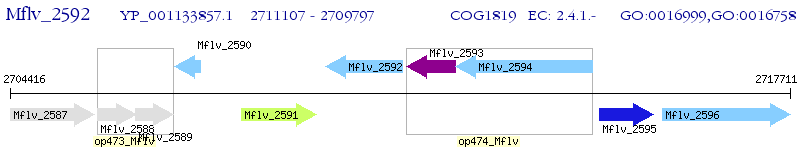
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2592 | - | - | 100% (436) | glycosyl transferase family protein |
| M. gilvum PYR-GCK | Mflv_1849 | - | 4e-82 | 42.58% (411) | UDP-glucuronosyl/UDP-glucosyltransferase |
| M. gilvum PYR-GCK | Mflv_0466 | - | 6e-35 | 29.93% (431) | glycosyl transferase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2986c | - | 4e-24 | 35.63% (247) | glycosyl transferase |
| M. tuberculosis H37Rv | Rv2962c | - | 5e-24 | 35.63% (247) | glycosyl transferase |
| M. leprae Br4923 | MLBr_00125 | - | 4e-24 | 35.05% (214) | putative glycosyl transferase |
| M. abscessus ATCC 19977 | MAB_3062c | - | 2e-19 | 27.21% (419) | hypothetical protein MAB_3062c |
| M. marinum M | MMAR_1755 | - | 2e-19 | 29.09% (220) | glycosyl transferase |
| M. avium 104 | MAV_3631 | - | 8e-17 | 29.33% (416) | hypothetical protein MAV_3631 |
| M. smegmatis MC2 155 | MSMEG_4670 | - | 1e-143 | 60.79% (431) | N-glycosylation |
| M. thermoresistible (build 8) | TH_3465 | - | 1e-80 | 40.78% (434) | PUTATIVE UDP-glucuronosyl/UDP-glucosyltransferase |
| M. ulcerans Agy99 | MUL_3383 | - | 6e-18 | 28.99% (414) | glycosyl transferase |
| M. vanbaalenii PYR-1 | Mvan_4034 | - | 0.0 | 73.66% (429) | glycosyl transferase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2592|M.gilvum_PYR-GCK ---------------------------------------MNAMPDILITA
Mvan_4034|M.vanbaalenii_PYR-1 ------------------------------------------MPEILIAT
MSMEG_4670|M.smegmatis_MC2_155 ------------------------------------------MADILIAS
TH_3465|M.thermoresistible__bu ------------------VGGYGLTRTRLLGRRLSSGSEDEFLARYLLAA
MAV_3631|M.avium_104 -------------------------------------------MRVAVVA
MUL_3383|M.ulcerans_Agy99 -------------------------------------------MRVAVVA
MAB_3062c|M.abscessus_ATCC_199 -------------------------------------------MRVAVVA
Mb2986c|M.bovis_AF2122/97 MRVSCVYATASRWGGPPVASEVRGDAAISTTPDAAPGLAARRRRILFVAE
Rv2962c|M.tuberculosis_H37Rv MRVSCVYATASRWGGPPVASEVRGDAAISTTPDAAPGLAARRRRILFVAE
MMAR_1755|M.marinum_M -------------------------MSIATN---AP-QPVRRQRVLFVAE
MLBr_00125|M.leprae_Br4923 ---------------------MGKDAGMSTAPDTVPECQSRRMRILFIAE
:.
Mflv_2592|M.gilvum_PYR-GCK CPPVGHIGPLLNVAHGLVAHGHRVTVLTSARHADKIRAVGADPRPLPYGA
Mvan_4034|M.vanbaalenii_PYR-1 CPPVGHLSPLLNVARGLVARGDRVTVLTSARHADKIRAVGAEPRPLPHAA
MSMEG_4670|M.smegmatis_MC2_155 HSPIGHIGPLLNVAYGLVGRGDRVTVLTSAQHTDAIRASGAAPVALPDAG
TH_3465|M.thermoresistible__bu TPLSGHVAPLVQIGATLRSRGHTVTVLTTTEFADAVTRAGLRFTALPAGT
MAV_3631|M.avium_104 GPDPGHSFPAIALCRRFADAGDTPTLFTGAEWLDTARGAGVDAVELDGLA
MUL_3383|M.ulcerans_Agy99 GPDPGHSFPAIALCRRFLAAGDIPTLLTGVEWLDTARAAGVDAIELAGLV
MAB_3062c|M.abscessus_ATCC_199 GPDPGHAFPALALCLKFAAAGDDPVLLTGTRWLDQARAAGVESVELLGLQ
Mb2986c|M.bovis_AF2122/97 AVTLAHVVRPFALAQSLDPSRYEVHFACDPRYNQLLGPLPFRHHAIHTIP
Rv2962c|M.tuberculosis_H37Rv AVTLAHVVRPFALAQSLDPSRYEVHFACDPRYNQLLGPLPFRHHAIHTIP
MMAR_1755|M.marinum_M ATTLAHVVRPFVVALSLDPSRYEVHFACDPRFNKLLGPIPFPHHPIYTIP
MLBr_00125|M.leprae_Br4923 AVTLAHVVRPFAVARLLDPSRYEVHFACDPRYNNLLGPLPFPHHPIHTVP
.* . : : . . . :
Mflv_2592|M.gilvum_PYR-GCK DYD----DARLDADLPGRAHTSGIARINFDVEHVFVRPMPYQFSAVQELL
Mvan_4034|M.vanbaalenii_PYR-1 DYD----DSTLDADLPGRAATAGLARINFDIESIFVRPLPHQFSALQELL
MSMEG_4670|M.smegmatis_MC2_155 DPD----ADRLDIENPKRARTSGIARINVDIVAFFVAPMAHQADALAKLL
TH_3465|M.thermoresistible__bu AIDPPAAPPWWARYLPSQARRFLLGRA--ELDSVYIRPLGHQAAALRSVL
MAV_3631|M.avium_104 A-----------------------TDEDVDAGARIHRRAARMAVLNVPAL
MUL_3383|M.ulcerans_Agy99 A-----------------------TDEDLDAGARIHRRAAQMALLNAPAL
MAB_3062c|M.abscessus_ATCC_199 PRV---------------------DDDDADSGAKIHQRAAHMALANLDQL
Mb2986c|M.bovis_AF2122/97 SER------------------FFGNLTQGR--FYAMRTLRKYVEADLRVL
Rv2962c|M.tuberculosis_H37Rv SER------------------FFGNLTQGR--FYAMRTLRKYVEADLRVL
MMAR_1755|M.marinum_M SER------------------FLGNLVQGR--FYTTRTLRQYVEEDSRVL
MLBr_00125|M.leprae_Br4923 SKR------------------FLDNMAQGRLFLYGPRTLRKYVEEDSKLL
*
Mflv_2592|M.gilvum_PYR-GCK AANSFDAVITDAFFLGVIPLLLDPTLARPPIMAYSTTPLFLSSRDTAPAG
Mvan_4034|M.vanbaalenii_PYR-1 VTDRFDAVIADALFLGTLPLLLGDPAKRPPILSYSTTPLFLTSRDTAPAG
MSMEG_4670|M.smegmatis_MC2_155 AQHRFDAVIADYGFFGILPFLLGNRADRPPFLTYSTTPLMLSSRDTAPAG
TH_3465|M.thermoresistible__bu DREPADAVLVDMTFTGAAALLLDDRP-RPPLLVCSVGPLTLSSADTPPFG
MAV_3631|M.avium_104 RDLAPDLVVSDVITAGGG---MAAELLGIPWIELSPHPLYLPSKGLPPIG
MUL_3383|M.ulcerans_Agy99 SDLAPDLVVSDVITACGG---MAAERLGIPWIELNPHPLYRPSKGLPPIG
MAB_3062c|M.abscessus_ATCC_199 RALAPDLIVSDVITACGG---MAGELLGVPWVELNPHPLYLPSKGLPPLG
Mb2986c|M.bovis_AF2122/97 DEIAPDLVVGDLRISLSVS----ARLAGIPYIAIAN--AYWSPYAQRRFP
Rv2962c|M.tuberculosis_H37Rv DEIAPDLVVGDLRISLSVS----ARLAGIPYIAIAN--AYWSPYAQRRFP
MMAR_1755|M.marinum_M AEVAPDLVVGDLRISLSAS----ARLAGIPYVAIAN--AYWSPYSRRRFP
MLBr_00125|M.leprae_Br4923 SEIEPDLVVGDLRWSLSVS----ARLASIPYIAIAN--AYWSSHARRRFP
* :: * * : ..
Mflv_2592|M.gilvum_PYR-GCK PGILPMPGAVGRLRNRLLGLLSRAVLLRPAHRSAERMLRAMGLPDLPVFV
Mvan_4034|M.vanbaalenii_PYR-1 PGIAPMPGMIGRLRNRVLARATQSVLLRPGQRAADRMLEAMGLPELPVFI
MSMEG_4670|M.smegmatis_MC2_155 LGLPPPRHAVERLRNRALETLTHKVLLRASQRAVQEQLHRMNCRPLPVFL
TH_3465|M.thermoresistible__bu MAWQPRPG----VDYRGMTAVAHRVIMAGSRRRFDRALRTAGAGPAPVFL
MAV_3631|M.avium_104 SGLATGTGLRGRLRDATMRALTARSWRAG--------LRQRAAARAEIGL
MUL_3383|M.ulcerans_Agy99 SGLAPGTGIRGRLRDTTMRALTARSWRAG--------LRQRAAVRAEIGL
MAB_3062c|M.abscessus_ATCC_199 SGLAPGVGLRGRLRDGLMRRLTQKSIDQG--------LRQRSQARVEIGL
Mb2986c|M.bovis_AF2122/97 LPDVIWTRLFGVRLVKLLYRLERPLLFALQCMPLNWVRRRHGLSSLGWNL
Rv2962c|M.tuberculosis_H37Rv LPDVIWTRLFGVRLVKLLYRLERPLLFALQCMPLNWVRRRHGLSSLGWNL
MMAR_1755|M.marinum_M LPEVFWSRLFGVRLVKRAYRLERPLFFALQCMPLNRVRRKHGLPSLGLNL
MLBr_00125|M.leprae_Br4923 LPDVLYTHLLGVRLVKLLYRLERPLFFAFQCLPLNWVRRKHGLPSLGFNL
. :
Mflv_2592|M.gilvum_PYR-GCK LECAILADRVIVPTVPDFEYPRRDLPAHVRFVGAVSPLPGDDFIAPAWWG
Mvan_4034|M.vanbaalenii_PYR-1 LDSAVLADRVIVPTVPEFEYRRSDLPSHVRFVGPVSPLPGSDFVAPPWWG
MSMEG_4670|M.smegmatis_MC2_155 FDGAVLSERVIAPTVPQFDYHRSDLPKNVRYVGAVHPMPNQGFRRPMWWR
TH_3465|M.thermoresistible__bu SDWPALADRLIQLGVPGFEYHRGDLPSTVDFAGPVLPTGLDGVTLPAWWD
MAV_3631|M.avium_104 PARDPGPLRRLIATLPALEVPRPDWPDEAVLVGPLHFEPTDRVLD-----
MUL_3383|M.ulcerans_Agy99 AEHDPGPLRRLIATLPALEVPRPDWPAEAIVVGPLHFEPTDQLLK-----
MAB_3062c|M.abscessus_ATCC_199 PARDPGPMRRLIATLPALEVPRPDWPAEAVVVGPLHWEPTDVVLQ-----
Mb2986c|M.bovis_AF2122/97 CRIFTDGDHTLYADVPELMP-TYDLPANHEYLGPVLWSPAGKPPT--WWD
Rv2962c|M.tuberculosis_H37Rv CRIFTDGDHTLYADVPELMP-TYDLPANHEYLGPVLWSPAGKPPT--WWD
MMAR_1755|M.marinum_M CRIFTDGDQTLYADIPELQP-TYNRPDNHQYLGPVLWSPSGETPD--WWD
MLBr_00125|M.leprae_Br4923 CRIFTDGDYTLYADVPELVP-TYDLPANHQYLGPVLWSPAGELPR--WWD
: :* : : * *.:
Mflv_2592|M.gilvum_PYR-GCK ELYRGRPVVHVTQGTVDNADLTRLIEPTIEALAG-EDVTVVVSTGGRPVS
Mvan_4034|M.vanbaalenii_PYR-1 ELNAGRPVVHVTQGTIDNADLTRLIEPTIEALAD-EDVTVVATTGGRPIS
MSMEG_4670|M.smegmatis_MC2_155 LLDGDRPVVHVTQGTVDNTDLTRLLEPTLEALAD-EDVVVIASTGGRDLD
TH_3465|M.thermoresistible__bu DIRSAATVVHVTQGTFDNTDLDQLVVPTLEALTDRPDVLVVATTGGRPGQ
MAV_3631|M.avium_104 -IPAGSGPVVVVAPSTAQTGARGLAEVALSCLTPGETLPAGARLVVSRLA
MUL_3383|M.ulcerans_Agy99 -VPAGSGPVLVIAPSTALSGTAGLAEVALESLIPGGTLSSGSRLVVSRLG
MAB_3062c|M.abscessus_ATCC_199 -PPSGSGPVVVIAPSTAFTGAEGMTELAVQALES-----AGARVVISALE
Mb2986c|M.bovis_AF2122/97 SLPTDRPIVYATLG---TSGGRNLLQLVLNALAELPVTVIAATAGRSDLK
Rv2962c|M.tuberculosis_H37Rv SLPTDRPIVYATLG---TSGGRNLLQLVLNALAELPVTVIAATAGRSDLK
MMAR_1755|M.marinum_M SLPADRPIVYATLG---TSGGRNLLQVVLNGLADLPVTVIAATADRSDLD
MLBr_00125|M.leprae_Br4923 SLPTDRPIVYATLG---TSGGKNLLQVVLDALADLPVTVIAATAGRSELQ
* . :. : .:. *
Mflv_2592|M.gilvum_PYR-GCK QIRTPLPANTFVAEFLPHDVLLPLVDVLVTNGGYGAVQRALAEGVPLVVA
Mvan_4034|M.vanbaalenii_PYR-1 QIRIPLPANTFVAKYVPHDVLLPMVDVMVTNGGYGAVQRALSDGVPVVVA
MSMEG_4670|M.smegmatis_MC2_155 VLRTPVPANTYVAKYIPHDLLMPKVDVLVTNGGYGTVQRALSCGVPLVVA
TH_3465|M.thermoresistible__bu RSGFRVPGNARVTGWLPYRMLMPHVDVMITNGGYGGVQYALSHGVPLIVA
MAV_3631|M.avium_104 GPELAAPP-WAVVGLGSQAELLRHADVVVCGGGHGMVAKTLLAGVPLVAV
MUL_3383|M.ulcerans_Agy99 GDDLVTPS-WAAVGLGSQAELLTHADVVICGGGHGMVSKTLLAGVPLVVV
MAB_3062c|M.abscessus_ATCC_199 PPSSGLPS-WAIAGLARQDLLLSGADLAVCGGGHGFLSKALLAGVPTVVV
Mb2986c|M.bovis_AF2122/97 T----VPANAFVADYLPGEAAAARSAVVVCNGGSLTTQQALVAGVPVIGV
Rv2962c|M.tuberculosis_H37Rv T----VPANAFVADYLPGEAAAARSAVVVCNGGSLTTQQALVAGVPVIGV
MMAR_1755|M.marinum_M D----IPANAYVADYLPGEAAAARSDVVICNGGSLTTQQALVAGVPVLGI
MLBr_00125|M.leprae_Br4923 N----VPANAFVADYLPGEAAAARSAVVICNGGSLTTQQAFVAGVPVVGI
* . : : .** :: *** :
Mflv_2592|M.gilvum_PYR-GCK GDTEDKPEVAARVEYFGVGVNLRTGTPAPQDLRDAVNAVLSDAGYRTRAG
Mvan_4034|M.vanbaalenii_PYR-1 GHTEDKPEVAARVRHFGVGIDLRTGTPTPTQVRRAVRKVLHEPGFRTKAG
MSMEG_4670|M.smegmatis_MC2_155 GKTEDKPEVAARVAWSGAGIDLRTGTPTPAAIRDAVREVLTDKRYLSRAR
TH_3465|M.thermoresistible__bu GETSDKAEIAARVEFCGAGIDLGTATPTVRALRAGFDRIRDQDRYRTAAR
MAV_3631|M.avium_104 PGGGDQWEIANRVVRQGSARLIRP--LSADALVAAVNEVLSSPGYRAAAQ
MUL_3383|M.ulcerans_Agy99 PGGGDQWEIANRVVRQGSARLIRP--LGADALAAAVNEVLSSPGYREAAQ
MAB_3062c|M.abscessus_ATCC_199 PGGGDQWELANRVVRQGSAELVRP--LSRDSLASAIGRVLGEPRYREAAR
Mb2986c|M.bovis_AF2122/97 AGNLDQHLNMEAVERAGAGVLLRTERLKSQRVAGAVMQVISRSEYRQAAA
Rv2962c|M.tuberculosis_H37Rv AGNLDQHLNMEAVERAGAGVLLRTERLKSQRVAGAVMQVISRSEYRQAAA
MMAR_1755|M.marinum_M ASNLDQHLNMQAIEAAGAGLLLRTERLKARSVSAAVCQLLDRPGYRQSAE
MLBr_00125|M.leprae_Br4923 AGNLDQHLNMEAVEAAGAGILLRSERLKVRRVADAVNRVLGQSEYRQAAQ
*: : * . : . : .. : : *
Mflv_2592|M.gilvum_PYR-GCK RLQAAYASRDSVTAIAALLDEVIPEQAAAR--------------------
Mvan_4034|M.vanbaalenii_PYR-1 WLRSAYAAYDSVAEIAKLVDEAVAQRTQPLQSA-----------------
MSMEG_4670|M.smegmatis_MC2_155 TLEAAFAQRDGVAEIIALVDEVIAERRAEVGGELACEVTHEQR-------
TH_3465|M.thermoresistible__bu RLAREIRSAAPLDSIAATLERIVAADALNRSARTGSAGAPDPDSTDDRPA
MAV_3631|M.avium_104 RAAAGIAD---VADPVRVCREALAG-------------------------
MUL_3383|M.ulcerans_Agy99 RAAASIAD---VADPVQVCHEALALAG-----------------------
MAB_3062c|M.abscessus_ATCC_199 RAAAGAIA---VEDPVRVCQDVLAALR-----------------------
Mb2986c|M.bovis_AF2122/97 RLADAFGRD--RVGFPQHVENALRLMPENRPRTWLAS-------------
Rv2962c|M.tuberculosis_H37Rv RLADAFGRD--RVGFPQHVENALRLMPENRPRTWLAS-------------
MMAR_1755|M.marinum_M ELAEVFGRE--LARFPQHVESALRRVSEGAVAKSAAG-------------
MLBr_00125|M.leprae_Br4923 RLAEVLERH--VAGLPQHMENVLHTVSQNPPPTSLARQKRPLRFG-----
:
Mflv_2592|M.gilvum_PYR-GCK ---------
Mvan_4034|M.vanbaalenii_PYR-1 ---------
MSMEG_4670|M.smegmatis_MC2_155 ---------
TH_3465|M.thermoresistible__bu PRPARPRVH
MAV_3631|M.avium_104 ---------
MUL_3383|M.ulcerans_Agy99 ---------
MAB_3062c|M.abscessus_ATCC_199 ---------
Mb2986c|M.bovis_AF2122/97 ---------
Rv2962c|M.tuberculosis_H37Rv ---------
MMAR_1755|M.marinum_M ---------
MLBr_00125|M.leprae_Br4923 ---------