For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TRPTLREISSLPAEPVRLADSALVLIDCQNTYTRGVMELEGVQAALDATAELLDRARTAGIPIIHIQHDS GPGSPYDVREEIGAIVESVAPRGDEPVVVKNFPNSFVQTDLQQRLQAVNASNLVLAGFMTHMCVNSTARG AFNLGFAPTVVAAATATRSLPGPDGNLVPASAMQSASLAAIADLFAVVVPDVGSIPD*
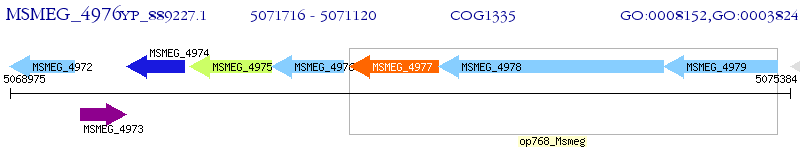
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_4976 | - | - | 100% (198) | isochorismatase hydrolase |
| M. smegmatis MC2 155 | MSMEG_4379 | - | 3e-07 | 25.88% (170) | isochorismatase hydrolase |
| M. smegmatis MC2 155 | MSMEG_4126 | - | 2e-05 | 28.36% (134) | hydrolase, isochorismatase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_2275 | - | 9e-78 | 75.13% (193) | isochorismatase hydrolase |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_3431c | - | 6e-09 | 27.91% (172) | isochorismatase hydrolase |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. thermoresistible (build 8) | TH_2198 | - | 5e-05 | 24.29% (177) | isochorismatase family protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_4420 | - | 3e-87 | 81.03% (195) | isochorismatase hydrolase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_2275|M.gilvum_PYR-GCK MSDPQTLRALAGLPLTPMSLADSALVLIDCQNTYTRGVMELDG-------
Mvan_4420|M.vanbaalenii_PYR-1 MSDLPTLRSLSGLPLQPVSLADSALVLIDCQNTYTYGVMELEG-------
MSMEG_4976|M.smegmatis_MC2_155 MTR-PTLREISSLPAEPVRLADSALVLIDCQNTYTRGVMELEG-------
TH_2198|M.thermoresistible__bu -------------------MSDTAVVVVDMFNTYDHEDAELLTP-----N
MAB_3431c|M.abscessus_ATCC_199 MTGQVDAQPYPWPFDGPVDPGRTAVLCIDWQVDFCGRGGYVDTMGYDLSL
. :*:: :* : :
Mflv_2275|M.gilvum_PYR-GCK VQAALDEAATLLDRARTAG-IAVVHIQHDDG--PGSLYDIE---------
Mvan_4420|M.vanbaalenii_PYR-1 VQAALDEAAALLDRARTAG-IPVVHIQHDDG--PGSPYDIT---------
MSMEG_4976|M.smegmatis_MC2_155 VQAALDATAELLDRARTAG-IPIIHIQHDSG--PGSPYDVR---------
TH_2198|M.thermoresistible__bu VESILDPLAKLIAEARHRDDVDLIYVNDNHGDFTAEFSDVVRT-------
MAB_3431c|M.abscessus_ATCC_199 TRAGLEPTARVLAAARAVG-MTIIHTREGHRPDLSDLPANKRWRSARIGA
..: *: * :: ** . : ::: ... ..
Mflv_2275|M.gilvum_PYR-GCK ---------------GESGAIVDRVAPRDGERVVVKNYPNSFVQTDLDDV
Mvan_4420|M.vanbaalenii_PYR-1 ---------------AEIGAIVDRVAPRGDEPVVVKNYPNSFVQTDLDDR
MSMEG_4976|M.smegmatis_MC2_155 ---------------EEIGAIVESVAPRGDEPVVVKNFPNSFVQTDLQQR
TH_2198|M.thermoresistible__bu ------------ALDGARPDLVRPIVPDDGCRALLKVRHSVFYATALEYL
MAB_3431c|M.abscessus_ATCC_199 EIGVAGPCGRILVKGEPGWEIVPEVAPLPGEPIIDKPGKGAFYATDLDLV
:* :.* . : * . * * *:
Mflv_2275|M.gilvum_PYR-GCK LKSYGASNLVLAGFMTHMCVNSTARGAFNLGYAPTVVAAATATRALPGVD
Mvan_4420|M.vanbaalenii_PYR-1 LKAVDASNLVLAGFMTHMCVNSTARGAFNLGYAPTVVAAATATRALPGVG
MSMEG_4976|M.smegmatis_MC2_155 LQAVNASNLVLAGFMTHMCVNSTARGAFNLGFAPTVVAAATATRSLPGPD
TH_2198|M.thermoresistible__bu LTRLSARRVILTGQVTEQCILYSALDAYVRHFEVVVPPDAVAH-------
MAB_3431c|M.abscessus_ATCC_199 LRTRGIRYIVLTGITTDVCVHTTMREANDRGFECLILSDCTGAT------
* . ::*:* *. *: : * : : . ...
Mflv_2275|M.gilvum_PYR-GCK GDTVAASAVHAASLAAVADLFAVVVPGEKDIPG-
Mvan_4420|M.vanbaalenii_PYR-1 GEDVPAAAVHSASLAAVADLFAVVVPGARDIPD-
MSMEG_4976|M.smegmatis_MC2_155 GNLVPASAMQSASLAAIADLFAVVVPDVGSIPD-
TH_2198|M.thermoresistible__bu ---IDAGLGEAALQMMARNMSAELPPSADCLP--
MAB_3431c|M.abscessus_ATCC_199 --DAGNHAAALKMVTMQGGVFGAVADSNQLLGAL
.: . : . :