For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SGFIETEEQQALRSAVAAMVAGYGQDYYLEKSRAGEHTDELWSEAGKLGFIGVNLPEEYGGGGAGMYELS LVMEEMAAGGASLLMMVVSPAINGTIIAKFGTEEQKKRWLPGIADGTLTMAFAITEPDAGSNSHRITTTA RRDGSDWILSGQKVYISGVDQAQAVLIVGRTEDHKTGNLKPALFVVPTDTPGLTYTKIPMEIVSPEFQFQ VFLDEVRLPADALVGSEDAAIAQLFAGLNPERIMGAASAVGMGRFALAKAVEYVKGRQVWKTPIGAHQGL SHPLAQNHIEVELAKLMMQKAAALYDAGADEAAAEAANMAKYAAGEASVRAVDQAVQSLGGNGLTKEYGV AAMLTASRLARIAPVSREMILNFVAQTSLGLPRSY*
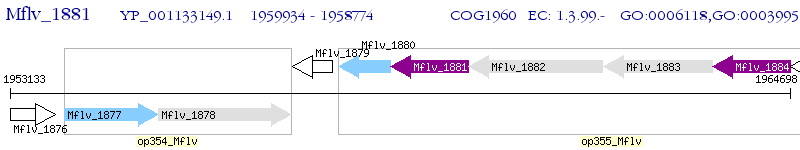
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1881 | - | - | 100% (386) | acyl-CoA dehydrogenase domain-containing protein |
| M. gilvum PYR-GCK | Mflv_4615 | - | e-129 | 59.84% (381) | acyl-CoA dehydrogenase domain-containing protein |
| M. gilvum PYR-GCK | Mflv_2442 | - | 2e-68 | 38.52% (392) | acyl-CoA dehydrogenase domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0997c | fadE12 | 0.0 | 82.60% (385) | acyl-CoA dehydrogenase FADE12 |
| M. tuberculosis H37Rv | Rv0972c | fadE12 | 0.0 | 82.86% (385) | acyl-CoA dehydrogenase FADE12 |
| M. leprae Br4923 | MLBr_00737 | fadE25 | 1e-34 | 28.06% (360) | putative acyl-CoA dehydrogenase |
| M. abscessus ATCC 19977 | MAB_1070c | - | 1e-160 | 72.06% (383) | acyl-CoA dehydrogenase |
| M. marinum M | MMAR_4535 | fadE12 | 0.0 | 83.38% (385) | acyl-CoA dehydrogenase FadE12 |
| M. avium 104 | MAV_1088 | - | 0.0 | 83.38% (385) | acyl-CoA dehydrogenase fadE12 |
| M. smegmatis MC2 155 | MSMEG_5494 | - | 0.0 | 88.60% (386) | acyl-CoA dehydrogenase fadE12 |
| M. thermoresistible (build 8) | TH_3553 | - | 0.0 | 83.29% (389) | PUTATIVE acyl-CoA dehydrogenase domain protein |
| M. ulcerans Agy99 | MUL_4707 | fadE12 | 0.0 | 83.12% (385) | acyl-CoA dehydrogenase FadE12 |
| M. vanbaalenii PYR-1 | Mvan_4850 | - | 0.0 | 92.69% (383) | acyl-CoA dehydrogenase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0997c|M.bovis_AF2122/97 -------MTDTSFIESEERQALRKAVASWVANYGHEYYLDKARKHEHTSE
Rv0972c|M.tuberculosis_H37Rv -------MTDTSFIESEERQALRKAVASWVANYGHEYYLDKARKHEHTSE
MMAR_4535|M.marinum_M -------MTDTSFIENEDRRALRKAVSEWASNYGHEYYLKKARAHEHTTE
MUL_4707|M.ulcerans_Agy99 ------MMTGTSFIENEDRRALRKAVSEWASNYGHEYYLKKARAHEHTTE
MAV_1088|M.avium_104 -------MTDSSFIENEERQALRKAVAEWASNYGSEYYLRKARAHEHTNE
Mflv_1881|M.gilvum_PYR-GCK ---------MSGFIETEEQQALRSAVAAMVAGYGQDYYLEKSRAGEHTDE
Mvan_4850|M.vanbaalenii_PYR-1 ---------MS-FIETDEQQALRKAVAAMAANYGQDYYLEKARAGQHTDE
MSMEG_5494|M.smegmatis_MC2_155 ---------MSGFVETEEQQALRRAVAAMAANYGQDYYLEKARAGQHTTE
TH_3553|M.thermoresistible__bu ---------VTGFVESEEQRALRRAAAELAAGYGQDYYLEKARAGEHTDE
MAB_1070c|M.abscessus_ATCC_199 ------MTQPNPYVESDERKALRQAVASLAASYGHEYYLEKSKTHEPLAE
MLBr_00737|M.leprae_Br4923 MVGWSGNPLFDLFKLPEEHNELRATIRALAEKEIAPHAADVDQRARFPEE
: :::. ** : . : : . *
Mb0997c|M.bovis_AF2122/97 LWAEAGKLGFLGVNLPEEYGGGGAGMYELSLVMEEMAAAGSALLLMVVSP
Rv0972c|M.tuberculosis_H37Rv LWAEAGKLGFLGVNLPEEYGGGGAGMYELSLVMEEMAAAGSALLLMVVSP
MMAR_4535|M.marinum_M LWSEAGKLGFIGVNLPEEYGGGGAGMYELSMVMEEMAAAGSALLLMVVSP
MUL_4707|M.ulcerans_Agy99 LWSEAGKLGFIGVNLPEEYGGGGAGMYELSMVMEEMAAAGSALLLMVVSP
MAV_1088|M.avium_104 LWSEAGKLGFLGVNLPEEYGGGGAGMYELSLVMEEMAAAGSALLLMVVSP
Mflv_1881|M.gilvum_PYR-GCK LWSEAGKLGFIGVNLPEEYGGGGAGMYELSLVMEEMAAGGASLLMMVVSP
Mvan_4850|M.vanbaalenii_PYR-1 LWSEAGKLGFIGVNLPEEYGGGGAGMYELSLVMEEMAAAGSSLLMMVVSP
MSMEG_5494|M.smegmatis_MC2_155 LWNEAGKLGFIGVNLPEEYGGGGAGMYELSLVMEEMAATGCALLMMVVSP
TH_3553|M.thermoresistible__bu LWAEAGKLGFLGVNLPEEYGGGGAGMYELSLVMEELSAAGCGLLMMVVSP
MAB_1070c|M.abscessus_ATCC_199 LWDEAGKLGFLGVNIPEEYGGGGAGMYELGLVFEELSAAGCPLLLMVVSP
MLBr_00737|M.leprae_Br4923 ALAALNASGFNAIHVPEEYGGQGADSVAACIVIEEVARVDASASLIPAVN
. ** .:::****** **. :*:**:: .. :: .
Mb0997c|M.bovis_AF2122/97 AINGTIIAKFGTDDQKKRWLPGIADGSLTMAFAITEPDAGSNSHKITTTA
Rv0972c|M.tuberculosis_H37Rv AINGTIIAKFGTDDQKKRWLPGIADGSLTMAFAITEPDAGSNSHKITTTA
MMAR_4535|M.marinum_M AINGTIISKFGTDEQKQRWLPGIADGTLTMAFAITEPDAGSNSHKITTTA
MUL_4707|M.ulcerans_Agy99 AINGTIISKFGTDEQKQRWLPGIADGTLTMAFAITEPEAGSNSHKITTTA
MAV_1088|M.avium_104 AINGTIISKFGTEEQKKRWLPGIADGTLTMAFAITEPDAGSNSHKITTTA
Mflv_1881|M.gilvum_PYR-GCK AINGTIIAKFGTEEQKKRWLPGIADGTLTMAFAITEPDAGSNSHRITTTA
Mvan_4850|M.vanbaalenii_PYR-1 AINGTIISKFGTEEQKKRWIPGIADGSLTMAFAITEPDAGSNSHKITTTA
MSMEG_5494|M.smegmatis_MC2_155 AINGTIISKFGTEEQKRRWLPGIADGSITMAFAITEPDAGSNSHRITTTA
TH_3553|M.thermoresistible__bu AINGTIISRFGTEDQKRRWLPGIADGSITMAFAITEPDAGSNSHRITTTA
MAB_1070c|M.abscessus_ATCC_199 AINATIIARYGTDAQKKSWLPAMAEGSFKMAFAITEPDAGSNSHNIITTA
MLBr_00737|M.leprae_Br4923 KLGTMGLILRGSEELKKQVLPSLAAEGAMASYALSEREAGSDAASMRTRA
:. : *:: *: :*.:* ::*::* :***:: : * *
Mb0997c|M.bovis_AF2122/97 RRDGSDWIIKGQKVFISGIDQAQAVLVVGRSEEAKTG---KLRPALFVVP
Rv0972c|M.tuberculosis_H37Rv RRDGSDWIIKGQKVFISGIDQAQAVLVVGRSEEAKTG---KLRPALFVVP
MMAR_4535|M.marinum_M RRDGSDWIIKGQKVYISGIDQAQAVLVVGRTEEAKTG---KLRPALFVVP
MUL_4707|M.ulcerans_Agy99 RRDGSDWIIKGQKVYISGIDQAQAVLVVGRTEEAKTG---KLRPALFVVP
MAV_1088|M.avium_104 RRDGSDWILKGQKVFISGIDQAQAVLVVARTEEAKTG---KLRPALFVVP
Mflv_1881|M.gilvum_PYR-GCK RRDGSDWILSGQKVYISGVDQAQAVLIVGRTEDHKTG---NLKPALFVVP
Mvan_4850|M.vanbaalenii_PYR-1 RRDGSDWVLSGQKVYISGVDQAQAVLVVGRTEDHKTG---SLKPALFVVP
MSMEG_5494|M.smegmatis_MC2_155 RRDGSDWILKGQKVFISGIDQAQAVLVVGRTEDHKTG---NLKPALFVIP
TH_3553|M.thermoresistible__bu RRDGGDWILSGQKVFISGVDQAQAVLVVGRVQKDGTGRTKSLKPALFVVP
MAB_1070c|M.abscessus_ATCC_199 KRDGGDWILNGRKVFISGVNEANAVLVVARTEEAKTG---KLKPALFVVP
MLBr_00737|M.leprae_Br4923 KADGDDWILNGFKCWITNGGKSTWYTVMAVTDPDKGAN----GISAFIVH
: **.**::.* * :*:. .:: ::. : . : *::
Mb0997c|M.bovis_AF2122/97 TDAPGFSYTPIEMEL-VSPERQFQVFLDDVRLPADALVGAEDAAIAHLFA
Rv0972c|M.tuberculosis_H37Rv TDAPGFSYTPIEMEL-VSPERQFQVFLDDVRLPADALVGAEDAAIAQLFA
MMAR_4535|M.marinum_M TDTPGFTYTAIDMEL-VSPERQFQVFLDDVRLPADALVGAQDAAIAQLFA
MUL_4707|M.ulcerans_Agy99 TDTPGFTYTAIDMEL-VSPERQFQVFLDDVRLPADALVGAQDAAIAQLFA
MAV_1088|M.avium_104 TDAPGFSATPIEMEL-ISPERQFQVFLDDVRLPSDALVGAEDAAIAQLFA
Mflv_1881|M.gilvum_PYR-GCK TDTPGLTYTKIPMEI-VSPEFQFQVFLDEVRLPADALVGSEDAAIAQLFA
Mvan_4850|M.vanbaalenii_PYR-1 TDTPGFTYTKIPMEI-VSPEFQFQVFLDEVRLPADALVGSEDAAIAQLFA
MSMEG_5494|M.smegmatis_MC2_155 TDTPGLHWTKIEMEL-ISPESQFQVFLDDVRVPADALVGSEDAALAQLFA
TH_3553|M.thermoresistible__bu TDAPGLSYTKIEMEL-ISPESQFQVFLDDVRLPADALVGDEDAALAQLFA
MAB_1070c|M.abscessus_ATCC_199 TNTKGFEYTPIDMQL-TLPEQQFQVFLDDVRVPGDALVGSEDAALMQLFA
MLBr_00737|M.leprae_Br4923 KDDEGFSIGPKEKKLGIKGSPTTELYFDKCRIPGDRIIGEPGTGFKTALA
.: *: :: . ::::*. *:*.* ::* .:.: :*
Mb0997c|M.bovis_AF2122/97 GLNPERIMGAASAVGMGRFALGRAVDYVKTRKVWSTPIGAHQGLAHPLAQ
Rv0972c|M.tuberculosis_H37Rv GLNPERIMGAASAVGMGRFALGRAVDYVKTRKVWSTPIGAHQGLAHPLAQ
MMAR_4535|M.marinum_M GLNPERIMGAASAVGMGRFALNRAVDYVKTRQVWSTPIGAHQGLAHPLAQ
MUL_4707|M.ulcerans_Agy99 GLNPERIMGAASAVGMGRFALNRAVDYVKTRQVWSTPIGAHQGLAHPLAQ
MAV_1088|M.avium_104 GLNPERIMGAASSVGMGRFALARAAEYVKTRQVWGTPVGAHQGLSHPLAQ
Mflv_1881|M.gilvum_PYR-GCK GLNPERIMGAASAVGMGRFALAKAVEYVKGRQVWKTPIGAHQGLSHPLAQ
Mvan_4850|M.vanbaalenii_PYR-1 GLNPERIMGAASAVGMGRFAIGRAVEYVKTRQVWKTPIGAHQGLSHPLAQ
MSMEG_5494|M.smegmatis_MC2_155 GLNPERIMGAASAVGMGRFAIHKAVDYVKTRQVWKVPIGSHQGLSHPLAQ
TH_3553|M.thermoresistible__bu GLNPERIMGAASAIGMGRFAIGRAVDYVKNRQVWRTPIGAHQGLAHPLAQ
MAB_1070c|M.abscessus_ATCC_199 GLNPERIMGAASGVGLTRLALNKATEYVKTRQVWKTPIGAHQAIAHPLAE
MLBr_00737|M.leprae_Br4923 TLDHTRPTIGAQAVGIAQGALDAAIVYTKDRKQFGESISTFQSIQFMLAD
*: * .*..:*: : *: * *.* *: : .:.:.*.: . **:
Mb0997c|M.bovis_AF2122/97 CHIEVELAKLMTQKAATLYDHGDDFGAAEAANMAKYAAAEASSRAVDQAV
Rv0972c|M.tuberculosis_H37Rv CHIEVELAKLMTQKAATLYDHGDDFGAAEAANMAKYAAAEASSRAVDQAV
MMAR_4535|M.marinum_M CHIEVELAKLMMQKAATLYDHGDDGGAAEAANMAKYAAAEASARSVDQAV
MUL_4707|M.ulcerans_Agy99 CHIEVELAKLMMQKAATLYDHGDDGGAAEAANMAKYAAAEASARSVDQAV
MAV_1088|M.avium_104 CHIEVELAKLMMQKAATLYDAGNDMGAAEAANMAKYAAAEAATRSVDQAV
Mflv_1881|M.gilvum_PYR-GCK NHIEVELAKLMMQKAAALYDAGADEAAAEAANMAKYAAGEASVRAVDQAV
Mvan_4850|M.vanbaalenii_PYR-1 NHIEVELAKLMMQKAASLYDAGDDAGAAEAANMAKYAAGEASVRAVDQAV
MSMEG_5494|M.smegmatis_MC2_155 NHIEIELAKLMMQKAAALYDAGDDLGAAEAANMAKYAAGEASVRAVDQAV
TH_3553|M.thermoresistible__bu NHIEIELAKLMMQKAAALYDLGDGAGAAEAANMAKYAAGEASARAVDQAV
MAB_1070c|M.abscessus_ATCC_199 IKVELELAKLMMQKAATLYDCGDDWGAAEAANMAKYAAADVACRAADRAV
MLBr_00737|M.leprae_Br4923 MAMKVEAARLIVYAAAARAERGEP-DLGFISAASKCFASDIAMEVTTDAV
:::* *:*: **: : * . : :* *.: : . . **
Mb0997c|M.bovis_AF2122/97 QSMGGNGLTKEYGVAAMMTSARLARIAPISREMVLNFVAQTSLGLPRSY
Rv0972c|M.tuberculosis_H37Rv QSMGGNGLTKEYGVAAMMTSARLARIAPISREMVLNFVAQTSLGLPRSY
MMAR_4535|M.marinum_M QSMGGNGLTQEYGVAAMLTSSRLARIAPISREMVLNFVAQTSLGLPRSY
MUL_4707|M.ulcerans_Agy99 QSMGGNGLTQEYGVAAMLTSSRLARIAPISREMVLNFVAQTSLGLPRSY
MAV_1088|M.avium_104 QSMGGNGLTKEYGVAAMLASARLGRIAPVSREMVLNFVAQTSLGLPRSY
Mflv_1881|M.gilvum_PYR-GCK QSLGGNGLTKEYGVAAMLTASRLARIAPVSREMILNFVAQTSLGLPRSY
Mvan_4850|M.vanbaalenii_PYR-1 QSLGGNGLTQEYGIASALTASRLARIAPVSREMILNFVAQTSLGLPRSY
MSMEG_5494|M.smegmatis_MC2_155 QSLGGNGLTKEYGIAAAVTASKLARIAPVSREMILNFVSQTSLGLPRSY
TH_3553|M.thermoresistible__bu QSLGGNGLTKEYGIAAAITSSRLARIAPISREMILNFVAQTSLGLPRSY
MAB_1070c|M.abscessus_ATCC_199 QSLGGNGLTTEYGVAPLLNLSRFGRIAPVSREMILNFVAQTSLGLPRSY
MLBr_00737|M.leprae_Br4923 QLFGGAGYTSDFPVERFMRDAKITQIYEGTNQIQRVVMSRALLR-----
* :** * * :: : : ::: :* :.:: .:::: *