For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VTSPNDLPRTVGELRASGHRERTVKAEIRENLLAALASGAPVDEVWPGILGFEDTVLPQLERALIAGHDI VLLGERGQGKTRLLRGLTGMLDEWTPVIAGAELGEHPYSPITPESIRRAADFGDDLPIAWRHRSERYTEK LATPDTSVADLVGDIDPIKVAEGRSLGDPETIAYGLIPRAHRGIVAVNELPDLAERIQVAMLNVMEERDI QVRGYTLRLPLDVLVVASANPEDYTNRGRIITPLKDRFGAEIRTHYPLELDAEVGVIVQEAHMAAEVPDY LLQILARFARALRESSSVDQRSGVSARFAIAAAETVAASARHRAAVLNEQEPVARVVDLASILDVLRGKL EFESGEEGREQAILEHLLRRATAETAQRLLGGIDVGPIVTAVEGGSPVTTGDRVSARDVLAALPEVSAVA EIQQRLGAQTDGQRAAAIELALEALYLAKRIDKVSDQGETVYG
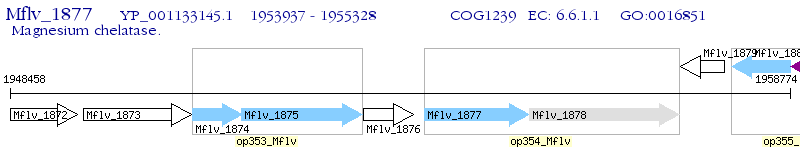
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1877 | - | - | 100% (463) | putative magnesium chelatase |
| M. gilvum PYR-GCK | Mflv_0039 | - | 2e-05 | 27.59% (116) | ATPase |
| M. gilvum PYR-GCK | Mflv_0476 | - | 8e-05 | 23.42% (269) | ATPase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0983 | - | 0.0 | 83.95% (461) | magnesium chelatase |
| M. tuberculosis H37Rv | Rv0958 | - | 0.0 | 83.73% (461) | magnesium chelatase |
| M. leprae Br4923 | MLBr_01810 | moxR | 2e-05 | 32.86% (70) | MoxR homolog |
| M. abscessus ATCC 19977 | MAB_0389c | - | 1e-05 | 25.95% (131) | putative regulatory protein |
| M. marinum M | MMAR_4541 | - | 0.0 | 84.02% (463) | magnesium chelatase |
| M. avium 104 | MAV_0412 | - | 2e-06 | 25.95% (131) | ATPase family protein associated with various cellular |
| M. smegmatis MC2 155 | MSMEG_5512 | - | 0.0 | 86.83% (463) | magnesium chelatase |
| M. thermoresistible (build 8) | TH_1334 | - | 0.0 | 86.61% (463) | POSSIBLE MAGNESIUM CHELATASE |
| M. ulcerans Agy99 | MUL_4711 | - | 0.0 | 84.23% (463) | magnesium chelatase |
| M. vanbaalenii PYR-1 | Mvan_4854 | - | 0.0 | 92.22% (463) | putative magnesium chelatase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0983|M.bovis_AF2122/97 --MSPSNLP----RTVGELRAAGHRERGVKQEIRENLLTALADGDN---V
Rv0958|M.tuberculosis_H37Rv --MSPSNLP----RTVGELRAAGHRERGVKQEIRENLLTALADGDN---V
MMAR_4541|M.marinum_M -MTSPSNLP----RTVGELRASGHRERGVKQEIRENLLTALADGPDNAAV
MUL_4711|M.ulcerans_Agy99 MVTSPSNLP----RTVGELRASGHRERGVKQEIRENLLTALADGPDNAAV
Mflv_1877|M.gilvum_PYR-GCK -MTSPNDLP----RTVGELRASGHRERTVKAEIRENLLAALASGAPVDEV
Mvan_4854|M.vanbaalenii_PYR-1 -MTSPNDLP----RTVGELRASGHQERSIKAEIRENLLSALASGAGADAV
MSMEG_5512|M.smegmatis_MC2_155 MVSQPKDLP----RTVGELRASGHRERGVKAEIRENLLAALAERR---DV
TH_1334|M.thermoresistible__bu -VTQPNDLP----RTVGELRASGHRERGVKQEIRENLLAALAGGR---DV
MAB_0389c|M.abscessus_ATCC_199 -MTQAT--------TTGSSGETARNALAALR-----------------EQ
MAV_0412|M.avium_104 -MTQPTQP-----VTASPTAESAREALLALR-----------------AE
MLBr_01810|M.leprae_Br4923 -MTSAGGLPVGADGCSGPGGQSGPRAHEAPPGGADGLVAEVHTLERAIFE
.. . :. .
Mb0983|M.bovis_AF2122/97 WPGILGFDDTVIPQVERALIAGHDFVLLGERGQGKTRLLRALAGLLDEWT
Rv0958|M.tuberculosis_H37Rv WPGILGFDDTVIPQVERALIAGHDFVLLGERGQGKTRLLRALAGLLDEWT
MMAR_4541|M.marinum_M WPGIVGFDDTVIPQLERALIAGHDFVLLGERGQGKTRLLRALTGLLDEWT
MUL_4711|M.ulcerans_Agy99 WPGIVGFDDTVIPQLERALIAGHDFVLLGERGQGKTRLLRALTGLLDEWT
Mflv_1877|M.gilvum_PYR-GCK WPGILGFEDTVLPQLERALIAGHDIVLLGERGQGKTRLLRGLTGMLDEWT
Mvan_4854|M.vanbaalenii_PYR-1 WPGILGFEDTVIPQLERALIAGHDVVLLGERGQGKTRLLRALTNLLDEWT
MSMEG_5512|M.smegmatis_MC2_155 WPGILGFEDTVIPQLERALIAGHDVVLLGERGQGKTRLLRALTGLLDEWT
TH_1334|M.thermoresistible__bu WPGILGFEDTVIPQLERALIAGHDVVLLGERGQGKTRLLRALIGLLDEWT
MAB_0389c|M.abscessus_ATCC_199 IAIVVVGQDSVVSGLVVALLCRGHVLLEGVPGVAKTLLVRTLS-------
MAV_0412|M.avium_104 LAKAVVGQEGVVSGLVIALLCRGHVLLEGVPGVAKTLLVRALA-------
MLBr_01810|M.leprae_Br4923 VKRVIVGQDQLVERMLVGLLSKGHVLLEGVPGVAKTLAVETFA-------
: :: :: : .*:. ..:* * * .** :. :
Mb0983|M.bovis_AF2122/97 PVIAGAELGEHPYTPITPESIRRAAQLGDDLPVAWKHRSERYTEKLATPD
Rv0958|M.tuberculosis_H37Rv PVIAGAELGEHPYTPITPESIRRAAQLGDDLPVAWKHRSERYTEKLATPD
MMAR_4541|M.marinum_M PVIAGAELPEHPYRPITPESIRRAAQLKDDLPVAWKHRSERYTEKLATPD
MUL_4711|M.ulcerans_Agy99 PVIAGAELPEHPYRPITPESIRRAAQLKDDLPVAWKHRSERYTEKLATPD
Mflv_1877|M.gilvum_PYR-GCK PVIAGAELGEHPYSPITPESIRRAADFGDDLPIAWRHRSERYTEKLATPD
Mvan_4854|M.vanbaalenii_PYR-1 PVIAGAELGEHPYSPITPESIRRAADSGDDLPITWRHRSERYTEKLATPD
MSMEG_5512|M.smegmatis_MC2_155 PVIAGSELGEHPYSPITPESIRRAADSGDDLPVEWRHRSERYTEKLATPD
TH_1334|M.thermoresistible__bu PVIAGSELGEHPYSPITPESIRRAADSGDDLPITWRHRSERYTEKLATPD
MAB_0389c|M.abscessus_ATCC_199 -----------------------------------AALSLDFKRVQFTPD
MAV_0412|M.avium_104 -----------------------------------AALQLEFKRVQFTPD
MLBr_01810|M.leprae_Br4923 -----------------------------------RVVGGTFARIQFTPD
: . ***
Mb0983|M.bovis_AF2122/97 TSVADLVGDVDPIKVAEGRSLGDPETIAYGLIPRAHRGIVAVNELPDLAE
Rv0958|M.tuberculosis_H37Rv TSVADLVGDVDPIKVAEGRSLGDPETIAYGLIPRAHRGIVAVNELPDLAE
MMAR_4541|M.marinum_M TSVADLVGDIDPIKVAEGRSLGDPETIAYGLIPRAHRGIVAVNELPDLAE
MUL_4711|M.ulcerans_Agy99 TSVADLVGDIDPIKVAEGRSLGDPETIAYGLIPRAHRGIVAVNELPDLAE
Mflv_1877|M.gilvum_PYR-GCK TSVADLVGDIDPIKVAEGRSLGDPETIAYGLIPRAHRGIVAVNELPDLAE
Mvan_4854|M.vanbaalenii_PYR-1 TSVADLVGDIDPIKVAEGRSLGDPETIAYGLIPRAHRGIVAVNELPDLAE
MSMEG_5512|M.smegmatis_MC2_155 TSVADLVGDIDPIKVAEGRSLGDPETIAYGLIPRAHRGIVAVNELPDLAE
TH_1334|M.thermoresistible__bu TSVADLVGDIDPIKVAEGRSLGDPETIAYGLIPRAHRGIVAVNELPDLAE
MAB_0389c|M.abscessus_ATCC_199 LMPGDVTGSLVYDARTAAFEFRAGPVFTN---------LLLADEINRTPP
MAV_0412|M.avium_104 LMPGDVTGSLVYDARTAAFVFRPGPVFTN---------LLLADEINRTPP
MLBr_01810|M.leprae_Br4923 LVPTDIIGTRIYRQGKEEFDTELGPVVVN---------FLLADEINRAPA
*: * ... :: .:*: .
Mb0983|M.bovis_AF2122/97 RIQVSMLNVMEERDIQVRGYTLRLPLDVLVVASANPEDYTNRGRIITPIK
Rv0958|M.tuberculosis_H37Rv RIQVSMLNVMEERDIQVRGYTLRLPLDVLVVASANPEDYTNRGRIITPIK
MMAR_4541|M.marinum_M RIQVSMLNVMEERDIQVRGYTLRLPLDVLVVASANPEDYTNRGRIITPLK
MUL_4711|M.ulcerans_Agy99 RIQVSMLNVMEERDIQVRGYTLRLPLDVLVVASANPEDYTNRGRIITPLK
Mflv_1877|M.gilvum_PYR-GCK RIQVAMLNVMEERDIQVRGYTLRLPLDVLVVASANPEDYTNRGRIITPLK
Mvan_4854|M.vanbaalenii_PYR-1 RIQVAMLNVMEERDIQVRGYTLRLPLDVLVVASANPEDYTNRGRIITPLK
MSMEG_5512|M.smegmatis_MC2_155 RIQVAMLNVMEERDIQVRGYTLRLPLDVLVVASANPEDYTNRGRIITPLK
TH_1334|M.thermoresistible__bu RIQVAMLNVMEERDIQVRGYTLRLPLDVLVVASANPEDYTNRGRIITPLK
MAB_0389c|M.abscessus_ATCC_199 KTQAALLEAMEERQVTVDGTPRPLPDPFIVAATQNPIEYEGTYQLPEAQL
MAV_0412|M.avium_104 KTQAALLEAMEERQVSVEGEAQPLPEPFIVAATQNPVEYEGTYQLPEAQL
MLBr_01810|M.leprae_Br4923 KVQSALLEVMQERHVSIGGKTFPLPNPFLVMATQNPIEHEGVYLLPEAQR
: * ::*:.*:**.: : * . ** .:* *: ** :: . : .
Mb0983|M.bovis_AF2122/97 DRFGAEIRTHYPLELEAEMGVIVQEAHLSAQVPDYLMQVLARFARYLRES
Rv0958|M.tuberculosis_H37Rv DRFGAEIRTHYPLELEAEMGVIVQEAHLSAQVSDYLMQVLARFARYLRES
MMAR_4541|M.marinum_M DRFGAEIRTHYPLELEAEMGVIAQEAHLSAQVPEYLLQVIARFARYLRES
MUL_4711|M.ulcerans_Agy99 DRFGAEIRTHYPLELEAEMGVIAQEAHLSAQVPDYLLQVIARFARYLRES
Mflv_1877|M.gilvum_PYR-GCK DRFGAEIRTHYPLELDAEVGVIVQEAHMAAEVPDYLLQILARFARALRES
Mvan_4854|M.vanbaalenii_PYR-1 DRFGAEIRTHYPPELDAEVGVITQEAHLAAEVPDYLLQILARFARSLRES
MSMEG_5512|M.smegmatis_MC2_155 DRFGAEIRTHYPLELEAEVGVIRQEAELAAEVPEYLLAVLARFARNLRES
TH_1334|M.thermoresistible__bu DRFGAEIRTHYPQQLEAEVGVITQEAHLAAEVPEYLLHILARFTRNLRES
MAB_0389c|M.abscessus_ATCC_199 DRFLMKLNVSLPERDQEIAILDRHAHGFDPRNLSHVTPVAS--AADLAAG
MAV_0412|M.avium_104 DRFLLKLNVTLPPREAEIAILHRHAHGFDPRDLSVVQPVAG--PADLAAG
MLBr_01810|M.leprae_Br4923 DRFLFKINVGYPSPEEEREIIYR--MGVTP---PVPKQILN--AGDLLRL
*** ::.. * : . . : . *
Mb0983|M.bovis_AF2122/97 RSIDQRSGVSARFAIAAAETVAAAARHRGAVLGETDPVARVVDLGTVIDV
Rv0958|M.tuberculosis_H37Rv RSIDQRSGVSARFAIAAAETVAAAARHRGAVLGETDPVARVVDLGTVIDV
MMAR_4541|M.marinum_M SSVDQRSGVSARFAIAAAETVAAAARHRGAVLGETDPVARVVDLSTVIDV
MUL_4711|M.ulcerans_Agy99 SSVDQRSGVSARFAIAAAETVAAAARHRGAVLGETDPVARVVDLGTVIDV
Mflv_1877|M.gilvum_PYR-GCK SSVDQRSGVSARFAIAAAETVAASARHRAAVLNEQEPVARVVDLASILDV
Mvan_4854|M.vanbaalenii_PYR-1 SSIDQRSGVSARFAIAAAETVAASARHRAALLGEQDPVARVVDLQNVIDV
MSMEG_5512|M.smegmatis_MC2_155 NSIDQRSGVSARFAIAAAETVAASARHRAAILGEDDPVARVVDLGTVIDV
TH_1334|M.thermoresistible__bu PSIDQRSGVSARFAIAAAETVAASARHRSAILGEDDPVARVVDLGTVIEV
MAB_0389c|M.abscessus_ATCC_199 RAAVGQVLVAPEVLAYVVDVIRATRQSPSL---------QLGASPRAATA
MAV_0412|M.avium_104 RDAVQQVRVADEVLGYIVDIVGATRSSPAL---------QLGVSPRGATA
MLBr_01810|M.leprae_Br4923 QEISANNFVHHALVDYVVRVVTATRHPEQLGMNDVKTWISFGASPRASLG
. * . . : *: .
Mb0983|M.bovis_AF2122/97 LRGKLEFESGEEGREQAVLEHLLRRATADTASRVLGGIDVGSLVTAVEGG
Rv0958|M.tuberculosis_H37Rv LRGKLEFESGEEGREQAVLEHLLRRATADTASRVLGGIDVGSLVTAVEGG
MMAR_4541|M.marinum_M LRGKLEFESGEEGREQAVLEHLLRRATADTASRALGGIDVGTLVSAVEGG
MUL_4711|M.ulcerans_Agy99 LRGKLEFESGEEGREQAVLEHLLRRATADTASRALGGIDVGTLVSAVEGG
Mflv_1877|M.gilvum_PYR-GCK LRGKLEFESGEEGREQAILEHLLRRATAETAQRLLGGIDVGPIVTAVEGG
Mvan_4854|M.vanbaalenii_PYR-1 LRGKLEFESGEEGREQAVLEHLLRRATAETAQRLLGGIDVGPIVTAVENG
MSMEG_5512|M.smegmatis_MC2_155 LRGKLEFETGEEGREQAVLEHLLRRATADTAQRLLGGIDVGPLVAAIEEG
TH_1334|M.thermoresistible__bu LRGKLEFESGEEGRELAILEHLLRRATADTAQRLLGGIDVGPLVAAVEEG
MAB_0389c|M.abscessus_ATCC_199 LLATSRAWAWLSGRNYVTPDDVKAMARPTLRHRIG--------LRPEAEL
MAV_0412|M.avium_104 LLATARSWSWLSGRNYVTPDDVKAMARPTLRHRVM--------LRPEAEL
MLBr_01810|M.leprae_Br4923 IIKASRSLALVRGRDYVIPQDVIEVIPDVLRHRLV--------LTYNALA
: . : **: . :.: * :
Mb0983|M.bovis_AF2122/97 SAVTTGERVSAKDVLAAVPGLPVVDRIARKLGAESEGERAAALELALEAL
Rv0958|M.tuberculosis_H37Rv SAVTTGERVSAKDVLAAVPGLPVVDRIARKLGAESEGERAAALELALEAL
MMAR_4541|M.marinum_M SAVTTGERVSAKDVLAAVPGLPVVETIARKLDAESEGERAAALELALEAL
MUL_4711|M.ulcerans_Agy99 SAVTTGERVSAKDVLAAVPGLPVVETIARKLNAESEGERAAALELALEAL
Mflv_1877|M.gilvum_PYR-GCK SPVTTGDRVSARDVLAALPEVSAVAEIQQRLGAQTDGQRAAAIELALEAL
Mvan_4854|M.vanbaalenii_PYR-1 SPVTTGERVSAREVLAAVPEVSAVAEIQQRLGATSEGQRAAAMELALEAL
MSMEG_5512|M.smegmatis_MC2_155 SPVTTGERVSAKDVVAALPDLPAIDAIAHRLNAVSVGQRAAAIELALEAL
TH_1334|M.thermoresistible__bu SPVTTGERVSAKDVLAALPDLPVIDTITTRLGAQSIGQRAAAVELALEAL
MAB_0389c|M.abscessus_ATCC_199 EGATSDRVLDGILVTVPVPR------------------------------
MAV_0412|M.avium_104 EGATCDGVLDGILASVPVPR------------------------------
MLBr_01810|M.leprae_Br4923 DEISPEIVINRVLQTVALPQVNSFPQQGHSVPSVTQAAAAASGR------
. : :. ..:*
Mb0983|M.bovis_AF2122/97 YLAKRVDKVCGEGQTVYG
Rv0958|M.tuberculosis_H37Rv YLAKRVDKVCGEGQTVYG
MMAR_4541|M.marinum_M YLAKRVDKVSGEGQTVYG
MUL_4711|M.ulcerans_Agy99 YLAKRVDKVSGEGQTVYG
Mflv_1877|M.gilvum_PYR-GCK YLAKRIDKVSDQGETVYG
Mvan_4854|M.vanbaalenii_PYR-1 YLAKRIDKVSDEGETVYG
MSMEG_5512|M.smegmatis_MC2_155 YLAKRIDKVSGEGETVYG
TH_1334|M.thermoresistible__bu YLAKRIDKQTGEGETIYG
MAB_0389c|M.abscessus_ATCC_199 ------------------
MAV_0412|M.avium_104 ------------------
MLBr_01810|M.leprae_Br4923 ------------------