For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SNTSAEQQDATAQSAWLSKSASETRGSDKKEVQFHYDISNDFFKLWQDPSQTYSCAYFEKDDYTLEQAQL AKVDLSLGKLGLQPGMTLLDIGCGWGSTIQRAVEKYDVNVIGLTLSENQKQHIEQNRFPNIDTNRSMEVR LQPWEEFDGKVDRIVSIGAFEHFGFNKYEDYFKKTFSWLPDDGVQMLHTIIIPSDEEIKAKKLPLTMSKV RFIKFIMDEIYPGGRLPLAAQVTDAAKGAGYTVTREQHLQPHYVKTLDTWAANLKDKKDEAVAITSEEIY ERFYKYLTGCADLFRDGYTDVVQFTLEKQ*
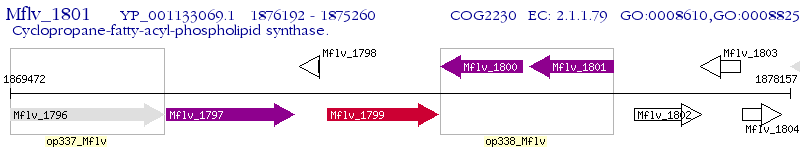
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1801 | - | - | 100% (310) | cyclopropane-fatty-acyl-phospholipid synthase |
| M. gilvum PYR-GCK | Mflv_3304 | - | e-104 | 62.95% (278) | cyclopropane-fatty-acyl-phospholipid synthase |
| M. gilvum PYR-GCK | Mflv_1800 | - | 7e-81 | 52.33% (279) | cyclopropane-fatty-acyl-phospholipid synthase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3424c | cmaA1 | 9e-98 | 60.29% (277) | cyclopropane-fatty-acyl-phospholipid synthase 1 CMAA1 |
| M. tuberculosis H37Rv | Rv3392c | cmaA1 | 1e-97 | 60.29% (277) | cyclopropane-fatty-acyl-phospholipid synthase 1 CMAA1 |
| M. leprae Br4923 | MLBr_02459 | umaA2 | 2e-97 | 61.07% (280) | Mycolic acid synthase |
| M. abscessus ATCC 19977 | MAB_3881 | - | 7e-94 | 58.63% (278) | cyclopropane-fatty-acyl-phospholipid synthase 1 |
| M. marinum M | MMAR_0979 | mmaA5_1 | 1e-101 | 60.79% (278) | methoxy mycolic acid synthase 5 MmaA5_1 |
| M. avium 104 | MAV_0130 | - | 1e-110 | 67.27% (278) | cyclopropane-fatty-acyl-phospholipid synthase 1 |
| M. smegmatis MC2 155 | MSMEG_1205 | - | 1e-123 | 73.08% (286) | cyclopropane-fatty-acyl-phospholipid synthase 1 |
| M. thermoresistible (build 8) | TH_2761 | - | 1e-115 | 68.95% (277) | PUTATIVE cyclopropane-fatty-acyl-phospholipid synthase 1 |
| M. ulcerans Agy99 | MUL_4539 | pcaA | 1e-100 | 62.82% (277) | mycolic acid synthase PcaA |
| M. vanbaalenii PYR-1 | Mvan_4948 | - | 1e-166 | 89.00% (309) | cyclopropane-fatty-acyl-phospholipid synthase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3424c|M.bovis_AF2122/97 -----------------MPD------ELKPHFANVQAHYDLSDDFFRLFL
Rv3392c|M.tuberculosis_H37Rv -----------------MPD------ELKPHFANVQAHYDLSDDFFRLFL
MMAR_0979|M.marinum_M -----------------MAK------ELKPHFDDVQAHYDLSDDFFRLFL
MLBr_02459|M.leprae_Br4923 -----------------MSV------QLTPHFRNVQAHYDLSDDFFRLFL
MUL_4539|M.ulcerans_Agy99 -----------------MSV------QLTPHFGNVQAHYDLSDDFFRLFL
MAB_3881|M.abscessus_ATCC_1997 -----------------MS-------KLTPKFSDVQAHYDLSDDFFALFL
MAV_0130|M.avium_104 -----------------MSENTGDAVNLQPHFDDVQAHYDLSDEFFALFL
Mflv_1801|M.gilvum_PYR-GCK MSNTSAEQQDATAQSAWLSKSASE--TRGSDKKEVQFHYDISNDFFKLWQ
Mvan_4948|M.vanbaalenii_PYR-1 MANSSTDQANKTAESAWLSKSAAQ--TRGSDKTEVQFHYDISNDFFKLWQ
MSMEG_1205|M.smegmatis_MC2_155 ----MKETPQKSNIAVKVGAPPGE--RHGSTADEVRSHYDLSNEFFRLWQ
TH_2761|M.thermoresistible__bu ----------------------------------VRSHYDLSNEFFRLWQ
*: ***:*::** *:
Mb3424c|M.bovis_AF2122/97 DPTQTYSCAYFERDDMTLQEAQIAKIDLALGKLGLQPGMTLLDVGCGWGA
Rv3392c|M.tuberculosis_H37Rv DPTQTYSCAYFERDDMTLQEAQIAKIDLALGKLGLQPGMTLLDVGCGWGA
MMAR_0979|M.marinum_M DPTQTYSCAYFERDDMTLEEAQIAKIDLALGKLGLEPGMTLLDVGCGWGA
MLBr_02459|M.leprae_Br4923 DPTLTYSCAYFERDDMTLEEAQLAKVDLALGKLKLEPGMTLLDIGCGWGA
MUL_4539|M.ulcerans_Agy99 DPSQTYSCAYFERDDMTLEQAQIAKIDLALGKLNLEPGMTLLDIGCGWGA
MAB_3881|M.abscessus_ATCC_1997 DPSRTYSCAYFEPETLTLEQAQQAKIDLALGKLNLEPGMTLLDVGCGWGS
MAV_0130|M.avium_104 DPTRTYSCAYFERDDMTLEEAQIAKVDLSLGKLGLQPGMTLLDIGCGWGT
Mflv_1801|M.gilvum_PYR-GCK DPSQTYSCAYFEKDDYTLEQAQLAKVDLSLGKLGLQPGMTLLDIGCGWGS
Mvan_4948|M.vanbaalenii_PYR-1 DPSQTYSCAYFERDDMTLEEAQLAKVDLSLGKLGLEPGMTLLDIGCGWGS
MSMEG_1205|M.smegmatis_MC2_155 DPTQTYSCAYFERDDMTLEEAQMAKVDLALGKLGLQPGMTLLDIGCGWGS
TH_2761|M.thermoresistible__bu DPTQTYSCAYFERDDMTLEEAQLAKVDLALGKLGLRPGMTLLDIGCGWGS
**: ******** : **::** **:**:**** *.*******:*****:
Mb3424c|M.bovis_AF2122/97 TMMRAVEKYDVNVVGLTLSKNQANHVQQL-VANSESLRSKRVLLAGWEQF
Rv3392c|M.tuberculosis_H37Rv TMMRAVEKYDVNVVGLTLSKNQANHVQQL-VANSENLRSKRVLLAGWEQF
MMAR_0979|M.marinum_M TMRRAIEKYDVNVVGLTLSKNQAAHVQKS-FDQLDTARTRRVLLEGWEQF
MLBr_02459|M.leprae_Br4923 TMKRAIEIYDVNVIGLTLSENQAAHVQKT-FDELDTPRTRRVLLEGWEKF
MUL_4539|M.ulcerans_Agy99 TMRRAIEKYDVNVVGLTLSENQAAHVQQM-FDQIDTSRSRRVLLEGWEKF
MAB_3881|M.abscessus_ATCC_1997 TMKRALEHYDVNVIGLTLSKNQFAYVNTL-LDSAGSSRNYEVRLAGWEEF
MAV_0130|M.avium_104 TIVRALERYDVNVVGLTLSRNQQAHVQRP-LDNHPSPRSKRVLLQGWEQF
Mflv_1801|M.gilvum_PYR-GCK TIQRAVEKYDVNVIGLTLSENQKQHIEQNRFPNIDTNRSMEVRLQPWEEF
Mvan_4948|M.vanbaalenii_PYR-1 TIARAVEKYDVNVIGLTLSENQKQHIEQNRFPQLKTDRKMEVRLQPWEEF
MSMEG_1205|M.smegmatis_MC2_155 TIMRAVEKYDVNVIGLTLSHNQRAHIEQR-FAESDSPRRKEVRLQPWEDF
TH_2761|M.thermoresistible__bu TIMRAVDEHDVDVIGLTLSENQRRHIEEHWFANSDSPRRKEVRLQGWEDF
*: **:: :**:*:*****.** ::: . . . * .* * **.*
Mb3424c|M.bovis_AF2122/97 DEPVDRIVSIGAFEHFGHERYDAFFSLAHRLLPADGVMLLHTITGLHPKE
Rv3392c|M.tuberculosis_H37Rv DEPVDRIVSIGAFEHFGHERYDAFFSLAHRLLPADGVMLLHTITGLHPKE
MMAR_0979|M.marinum_M DEPVDRIVSIGAFEHFGHDRYDDFFTLAHNILPSDGVMLLHTITGLTFQQ
MLBr_02459|M.leprae_Br4923 HEPVDRIVSIGAFEHFGRQRYARFFKMAHEVLPADSVMLLHTIVRPTFKD
MUL_4539|M.ulcerans_Agy99 DEPVDRIVSIGAFEHFGRLRYAHFFKMAYQALPADGIMLLHTIARPTFKE
MAB_3881|M.abscessus_ATCC_1997 EGTVDRIVSIGAFEHFGHERYDSFFAMAHRVLPSDGKMLLHTIVSHHPDE
MAV_0130|M.avium_104 DEKVDRIVSIGAFEHFGRDRYDDFFKMAYAALPDDGVMMLHTIIKPSDEE
Mflv_1801|M.gilvum_PYR-GCK DGKVDRIVSIGAFEHFGFNKYEDYFKKTFSWLPDDGVQMLHTIIIPSDEE
Mvan_4948|M.vanbaalenii_PYR-1 DGKVDRVVSIGAFEHFGFNKYDDYFKKTFSWMPDDGVMLLHTIIIPSDEE
MSMEG_1205|M.smegmatis_MC2_155 DEPVDRIVSIGAFEHFGFEKYADYFKKTFELMPDDGVMLLHTIVSTSKEE
TH_2761|M.thermoresistible__bu DEPVDRIVSIGAFEHFGFDNYDFYFKKTYDLLPDDGVMLLHTIVSTSREE
. ***:********** .* :* :. :* *. :**** .:
Mb3424c|M.bovis_AF2122/97 IHERGLPMSFTFARFLKFIVTEIFPGGRLPSIPMVQECASANGFTVTRVQ
Rv3392c|M.tuberculosis_H37Rv IHERGLPMSFTFARFLKFIVTEIFPGGRLPSIPMVQECASANGFTVTRVQ
MMAR_0979|M.marinum_M ATDRGMPLTFEIARFIKFIVTEIFPGGRLPSIEKVADHSSKAGFTLTRRQ
MLBr_02459|M.leprae_Br4923 ARARGMTLTHEIVHFTQFILAEIFPGGWLPTVPTVEEHATAAGFKFTRIQ
MUL_4539|M.ulcerans_Agy99 ARAKGLTLTHEIVHFSQFILAEIFPGGWLPTIPTVEEHATAAGFKVTRVQ
MAB_3881|M.abscessus_ATCC_1997 WKRMGIPLLMSRVRFIYFIGTEIFPGGRLPSVPMVDKHAALAGFHITRHH
MAV_0130|M.avium_104 FAERGLPITMTKLRFMKFIMDEIFPGGDLPQAKGVIEHAQRAGFGVQRVQ
Mflv_1801|M.gilvum_PYR-GCK IKAKKLPLTMSKVRFIKFIMDEIYPGGRLPLAAQVTDAAKGAGYTVTREQ
Mvan_4948|M.vanbaalenii_PYR-1 IKAKKLPLTMSNVRFIKFIMDEIYPGGRLPLAAQVTDAATRNGYTVTREQ
MSMEG_1205|M.smegmatis_MC2_155 VEEMGLPTTMSLMRFFRFIVTEIYPGGRIPLIAMVEDRATEAGFTVTRKQ
TH_2761|M.thermoresistible__bu VAELGLPTTMSLLRFFKFIVDEIYPGGRIPLIAQVEDAATNAGFRITQKQ
:. :* ** **:*** :* * . : *: . : :
Mb3424c|M.bovis_AF2122/97 SLQPHYAKTLDLWSAALQANKGQAIALQSEEVYERYMKYLTGCAEMFRIG
Rv3392c|M.tuberculosis_H37Rv SLQPHYAKTLDLWSAALQANKGQAIALQSEEVYERYMKYLTGCAEMFRIG
MMAR_0979|M.marinum_M SLQPHYARTLDLWAQALQARRDEAIEVQSEEVYERYMKYLTGCADGFRVG
MLBr_02459|M.leprae_Br4923 SLQLHYAKTLDLWAARLEANKNKAIAIQSEEVYDRYMKYLTGCAKLFRDG
MUL_4539|M.ulcerans_Agy99 SLQLHYARTLEIWAAALEAKKEEAIAIQSQKVYDRYMKYLTGCAKLFRQG
MAB_3881|M.abscessus_ATCC_1997 SLQPHYARTLDCWATNLAAHEDEAISVQSQEVYDRYIKYLTGCAEGFREG
MAV_0130|M.avium_104 ALRPHYARTLDTWAAALRARRDEAIAIQSEEVYDRYMKYLTGCADLFREG
Mflv_1801|M.gilvum_PYR-GCK HLQPHYVKTLDTWAANLKDKKDEAVAITSEEIYERFYKYLTGCADLFRDG
Mvan_4948|M.vanbaalenii_PYR-1 HLQPHYVKTLDTWASNLKDKKDEAIAITSEQIYERFYKYLTGCADLFRDG
MSMEG_1205|M.smegmatis_MC2_155 RLRPHYVRTLDTWAANLEAKKDQAIEITSEEVYERYLKYLKGCADLFRDG
TH_2761|M.thermoresistible__bu RLRPHYVKTLDAWAANLEANRRAAIEITSEEVYDRYLRYLTGCSDLFAKG
*: **.:**: *: * .. *: : *:::*:*: :**.**:. * *
Mb3424c|M.bovis_AF2122/97 YIDVNQFTCQK--------
Rv3392c|M.tuberculosis_H37Rv YIDVNQFTCQK--------
MMAR_0979|M.marinum_M YIDVNQFTLEKS-------
MLBr_02459|M.leprae_Br4923 YTDVDQFTLEKQPATARRH
MUL_4539|M.ulcerans_Agy99 YTDIDQFTLEK--------
MAB_3881|M.abscessus_ATCC_1997 WIDVVQFTCEK--------
MAV_0130|M.avium_104 YTDVAQFTLTKG-------
Mflv_1801|M.gilvum_PYR-GCK YTDVVQFTLEKQ-------
Mvan_4948|M.vanbaalenii_PYR-1 YTDVVQFTCEKSGV-----
MSMEG_1205|M.smegmatis_MC2_155 YTDVCQFTCEKSVA-----
TH_2761|M.thermoresistible__bu YTDVCQFTCRKGQ------
: *: *** *