For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VAIDRSIGETQRWAYPLLLVLSGVALGVSGLPAPLYGIYETNWHLSPLATTIVFAVYAVAALAAVLVSGR ISDVVGRKPVLLGALVALLIGLGVFLIADSMAMLLLARAIHGVAVGSIVVAGAAALLDLRPDHGVRVGQL SGVAFNIGMTIAIFGSALLAQYAPHPLRTPYAVVAVICLIVGVGVLALREPHTARTRGPIRIARPAVPAE IRADFWFSALGAMASWSVLGVLLSLYPSLAAHHTHIDNLVFGGAVVATTAFAAALAQLAATRVPARYAAI IGDAGMAAALLLTIPVLLTHQWQLVFVAAALLGATFGLGFGGSLRHLSNVVPPARRGETMSAFYLLAYSA MAVPTLVAGWAATRWELASVFPWFAGIVSAACLGAAVAGLRSIRVARAA*
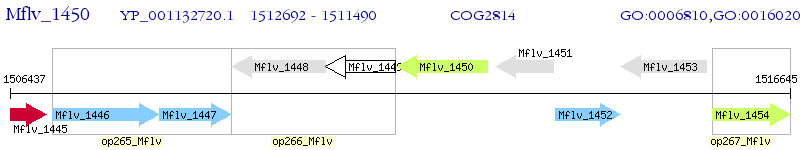
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1450 | - | - | 100% (400) | major facilitator transporter |
| M. gilvum PYR-GCK | Mflv_1534 | - | 2e-22 | 27.30% (381) | major facilitator transporter |
| M. gilvum PYR-GCK | Mflv_2728 | - | 9e-11 | 21.45% (387) | major facilitator transporter |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1908 | - | 3e-11 | 25.00% (228) | hypothetical protein Mb1908 |
| M. tuberculosis H37Rv | Rv1877 | - | 1e-13 | 24.01% (304) | integral membrane protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_1299c | - | 6e-43 | 30.40% (375) | multidrug ABC transporter |
| M. marinum M | MMAR_0690 | - | 1e-10 | 26.25% (339) | putative transport protein |
| M. avium 104 | MAV_0138 | - | 2e-21 | 27.53% (396) | transporter, major facilitator family protein |
| M. smegmatis MC2 155 | MSMEG_5167 | - | 7e-52 | 30.98% (397) | major facilitator superfamily protein MFS_1 |
| M. thermoresistible (build 8) | TH_0476 | - | 7e-47 | 29.82% (379) | major facilitator superfamily protein MFS_1 |
| M. ulcerans Agy99 | MUL_3826 | - | 6e-07 | 29.41% (153) | hypothetical protein MUL_3826 |
| M. vanbaalenii PYR-1 | Mvan_5329 | - | 0.0 | 92.21% (398) | major facilitator superfamily transporter |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1450|M.gilvum_PYR-GCK ----------------MVAIDRSIGETQR-WAYPLLLVLSGVALGVSGLP
Mvan_5329|M.vanbaalenii_PYR-1 ----------------MVAFDRSIPDTQR-WAYPLLLVLSGVALGVSGLP
MSMEG_5167|M.smegmatis_MC2_155 -----------------------MPHTRR-FSFALLAYAFAAIMVGTTLP
TH_0476|M.thermoresistible__bu --------------------------VRR-FAFPLLAYAFAAVMVGTTVP
MAB_1299c|M.abscessus_ATCC_199 ----------------------MIARLRRSAAFVLLAFSFSAVMIGTTMP
Rv1877|M.tuberculosis_H37Rv MAGP---TAPTTAPTAIRAGGPLLSPVRRNIIFTALVFGVLVAATGQTIV
MUL_3826|M.ulcerans_Agy99 MMRPRATVAAATPTTDLAAGRALITPRRRNFIFVALVLGILLSSLDQTIV
Mb1908|M.bovis_AF2122/97 MAGP---TAPTTAPTAIRAGGPLLSPVRRNIIFTALVFGVLVAATGQTIV
MAV_0138|M.avium_104 ------MMSSNVRAARNRPGRTDPQPPTGAPLFVGVLGLLLATGWVANHF
MMAR_0690|M.marinum_M -----------------------MTDSSRRVVLAAVGFTFFIDMLVYGIV
:
Mflv_1450|M.gilvum_PYR-GCK APLYGIYETNWHLSPLATTIVFAVYAVAALAAVLVSGRISDVVGRKPVLL
Mvan_5329|M.vanbaalenii_PYR-1 APLYGIYETNWHLSPLATTVVFAVYAVAALAAVLVSGRISDVVGRKPVLV
MSMEG_5167|M.smegmatis_MC2_155 TPMYALYAEHMNFAVLTTTVIYATYAGGVLFALLLFGRWSDAIGRRPMLL
TH_0476|M.thermoresistible__bu TPMYAVYAERLGFGVLTTTVVFAGYTAGVLFALVVFGRWSDALGRRPMLL
MAB_1299c|M.abscessus_ATCC_199 TPMYALYSQQMHFSVLTTTVVYATYAAGVLFALLMFGGWSDVLGRRPLLL
Rv1877|M.tuberculosis_H37Rv VPALPTIVAELG-STVDQSWAVTSYLLGGTVVVVVAGKLGDLLGRNRVLL
MUL_3826|M.ulcerans_Agy99 AIALPTIVADLG-EAGRQSWVVTSYLLASTIATALVGKLGDMFGRKRVFQ
Mb1908|M.bovis_AF2122/97 VPALPTIVAELG-STVDQSWAVTSYLLGGTVVVVVAGKLGDLLGRNRVLL
MAV_0138|M.avium_104 VGLMPAISDRDHLATTTLDGIFGIYALGLLPGLLVGGRTSDALGRRPVAL
MMAR_0690|M.marinum_M VPVLPRLAKPAGLEFLGVGVVFACYGLAYFVLTPVAGWLIDRRGSRSVFL
. * . : * * * . :
Mflv_1450|M.gilvum_PYR-GCK GALVALLIGLGVFLIADSMAMLLLARAIHGVAVGSIVVAGAAALLDLRPD
Mvan_5329|M.vanbaalenii_PYR-1 GALVALLIGLGVFLIADSMAMLLLARTIHGAAVGSIVVAGAAALLDLRPD
MSMEG_5167|M.smegmatis_MC2_155 AGVAAALISAGVFLMADSVPVLLIGRIFSGLSAGIFTGTATAAVIEAAPP
TH_0476|M.thermoresistible__bu AGVGCAIVSALVFLIAGSVPQLIAARFVSGMSAGIFTGTATAAVIEAAPE
MAB_1299c|M.abscessus_ATCC_199 TGAGFAILSSVIFLFVDSVPLLLVARIVSGLSAGFFTGAATVAVIESAPE
Rv1877|M.tuberculosis_H37Rv GSVVVFVVGSVLCGLSQTMTMLAISRALQGVGAGAISVTAYALAAEVVPL
MUL_3826|M.ulcerans_Agy99 VAVLLFVAGSVSCGLTQSMTMLVASRALQGVGGGAITVTAIALIGEVVPL
Mb1908|M.bovis_AF2122/97 GSVVVFVVGSVLCGLSQTMTMLAISRALQGVGAGAISVTAYALAAEVVPL
MAV_0138|M.avium_104 TGSAAALVGTVAMLLSQHSPALFAGRLIVGLGVGLAISAGTAWASDLRGP
MMAR_0690|M.marinum_M SGAIILMLATLAFGYASSPVGLVTCRVLQGGAAAATWVAGYATLVEIFPR
. : . * * . * . . :. . :
Mflv_1450|M.gilvum_PYR-GCK HG-VRVGQLSGVAFNIGMTIAIFGSALLAQYAPH--PLRTPYAVVAVICL
Mvan_5329|M.vanbaalenii_PYR-1 HG-VRAGQLSGVSFNIGMTVAILGSALLAQYAPH--PLRTPYAVVAVICL
MSMEG_5167|M.smegmatis_MC2_155 AWRTRAAAVATVANIGGLGTGPLLAGLLVEYAPR--PLHLSFVVHIVMAV
TH_0476|M.thermoresistible__bu RRRSRAAAVATVANIGGLGAGPLLAGILVQYAPW--PLHLTFWVHIVLMV
MAB_1299c|M.abscessus_ATCC_199 TWRGRAAAVATVANTGGLGLGPLLAGVLVQYAPW--PTHLAFVVHIGLVS
Rv1877|M.tuberculosis_H37Rv RDRGRYQGVLGAVFGVNTVTGPLLGGWLTDYLSWRWAFWINVPVSIAVLT
MUL_3826|M.ulcerans_Agy99 RDRGRYQGILGAVIGIATIGGPLLGGYFTDCLGWRWAFWINLPVSAVVIC
Mb1908|M.bovis_AF2122/97 RDRGRYQGVLGAVFGVNTVTGPLLGGWLTDYLSWRWAFWINVPVSIAVLT
MAV_0138|M.avium_104 AG----AATAGAVLTAGFAVGPFAGGVLAWAGPS--GVRASFALAAAILA
MMAR_0690|M.marinum_M EQRGRAVGLASIGTALGALLGPALGGLAYEASGP----HAPFLGAAGLSA
. .. :
Mflv_1450|M.gilvum_PYR-GCK IVGVGVLALRE-PHTARTRGPIRIARPAVPAEIRADFWFSALGAMASWSV
Mvan_5329|M.vanbaalenii_PYR-1 VVGVGVLALRE-PHTARTRGPIRIARPAVPAEIRADFWFSALGAMASWSV
MSMEG_5167|M.smegmatis_MC2_155 LAGAAVFAV---PETSSRSGSIGLQRLSVPPEVRTVFVIAAIAAFAGFAV
TH_0476|M.thermoresistible__bu LAAAAIAVA---PETSPRTGRLGVQRLSLPPQVRKVFAIASLAAFAGFAV
MAB_1299c|M.abscessus_ATCC_199 IAVAALLFV---PETAPKHGKLGRQRLSLPPQVRATFAAAATAGFAGFAA
Rv1877|M.tuberculosis_H37Rv VAATAVPALARPPKPVIDYLGILVIAVATTALIMATSWGGTTYAWGSATI
MUL_3826|M.ulcerans_Agy99 VATAAIPALAATTRPVIDYAGIMFIGLSLAALTLATSLGGSVYAWGSAPI
Mb1908|M.bovis_AF2122/97 VAATAVPALARPPKPVIDYLGILVIAVATTALIMATSWGGTTYAWGSATI
MAV_0138|M.avium_104 LAAFAVVAAPQ-PSPVTAPADPDGEETADAAPQGISRALSWAMPLAPWVF
MMAR_0690|M.marinum_M LAVAALAASLPQAAGGQASGPGMWSGERAFPGLAVPWLVAGLAVGLALGA
:. .: . . .
Mflv_1450|M.gilvum_PYR-GCK LGVLLSLYPSLAAHHTHIDNLVFGGAVVATTAFAA---------------
Mvan_5329|M.vanbaalenii_PYR-1 LGVLLSLYPSLAARQTHIDNLVFGGAVVGTTAFAA---------------
MSMEG_5167|M.smegmatis_MC2_155 TGMYTAVAPSFLSNVIGVDNHAVAGAVACSIFAAS---------------
TH_0476|M.thermoresistible__bu TGLFTAVAPSFLAEVIGVRNHAVAGAVAASIFAAS---------------
MAB_1299c|M.abscessus_ATCC_199 LGSFTAVAPGFLSGVLGIDNHAVAGASVFLTFGSS---------------
Rv1877|M.tuberculosis_H37Rv VGLLIGAA-VALGFFVWLEGRAAAAILPPRLFGSPVFAVCCVLSFVVGFA
MUL_3826|M.ulcerans_Agy99 IGLFTAAG-VTLAVFVWVETIAAQPILPIRLFAAPVFSVCCVLAFVVGFA
Mb1908|M.bovis_AF2122/97 VGLLIGAA-VALGFFVWLEGRARCGHPAAQAVWQP--SICRVLR------
MAV_0138|M.avium_104 ASATLGFVTIPGRLHTALAAPVAAGTATLIVNGVS---------------
MMAR_0690|M.marinum_M VEATLPIDLVARFDASGVTVGLLFALTTVAFIVTS---------------
: .
Mflv_1450|M.gilvum_PYR-GCK --------------------------------------ALAQLAATRVPA
Mvan_5329|M.vanbaalenii_PYR-1 --------------------------------------ALAQLAATRVPA
MSMEG_5167|M.smegmatis_MC2_155 --------------------------------------AVTQVLANRVQP
TH_0476|M.thermoresistible__bu --------------------------------------AVTQVLGASVSP
MAB_1299c|M.abscessus_ATCC_199 --------------------------------------CVAQILTRRVAA
Rv1877|M.tuberculosis_H37Rv MLGALTFVPIYLGYVDGASATASGLRTLPMVIGLLIASTGTGVLVGRTGR
MUL_3826|M.ulcerans_Agy99 MLGALIFVPTFMQYVNGVSATASGLRILPMVIGMLITSIGSGSMVGRTGR
Mb1908|M.bovis_AF2122/97 --------PVLRGRIRDAGCTD-------------LRTDLSGVRGRRVG-
MAV_0138|M.avium_104 --------------------------------------GAVQVLARALRW
MMAR_0690|M.marinum_M -----------------------------------------QYAGRLSDA
Mflv_1450|M.gilvum_PYR-GCK RYAAIIGDAGMAAALLLTIPVLLTHQWQLV-FVAAALLGATFGLGFG---
Mvan_5329|M.vanbaalenii_PYR-1 RYSAILGDAGMAAALLLTIPVLLTHQWQLV-FVAAALLGATFGLGFG---
MSMEG_5167|M.smegmatis_MC2_155 GRAVAVGCAILIAGMLILTAALLFSSLPAL-IAAAVVSGVGQGMSFS---
TH_0476|M.thermoresistible__bu RRAVAVGCALLIVGMVFLAAALVWSSLPAL-IATAVVAGAGQGLSFS---
MAB_1299c|M.abscessus_ATCC_199 AKALLIGCTILLTGMLLVVLALHTSSLPWY-LASAVVVGIGQGISFS---
Rv1877|M.tuberculosis_H37Rv YKIFPVAGMALMAVAFLLMSQMD-EWTPPL-LQSLYLVVLGAGIGLS-MQ
MUL_3826|M.ulcerans_Agy99 YKIFPVLGTALMTLAFLLMSRTD-ESTSAA-VQSVYLLILGSAIGMS-SQ
Mb1908|M.bovis_AF2122/97 ----DRVRSAHVADGDRPADRLDRDGCPGR-PDGPLQDLPGRGDGADGGC
MAV_0138|M.avium_104 GPQAGTAGAVLAALGYAVAAAAPSTLTPALGVPLFVVLGCASGLCLR---
MMAR_0690|M.marinum_M GRRAPVLAAGWVTLG-VALAVMGVPSAVGIQAAALVLLGAGLGAMIA---
. .
Mflv_1450|M.gilvum_PYR-GCK --------------GSLRHLSNVVPPARRGETMSAFYLLAYSAMAVPTLV
Mvan_5329|M.vanbaalenii_PYR-1 --------------GSLRHLSDVVPAGRRGETMSAFYLLAYSAMAVPTLI
MSMEG_5167|M.smegmatis_MC2_155 --------------RGLAAIAEKTPPDRRAEVSSTYFVVAYVAISLPVVG
TH_0476|M.thermoresistible__bu --------------RGLAAVSARTPSDQRGEVSSTYFVVAYVALLVPVIG
MAB_1299c|M.abscessus_ATCC_199 --------------RGLAAIADRTPEDNRAEVTSSYFVVAYIGIALPVIG
Rv1877|M.tuberculosis_H37Rv VLVLIVQNTSSFEDLGVATSGVTFFRVVGASFGTATFGALFVNFLDRRLG
MUL_3826|M.ulcerans_Agy99 VLVIIVQNTSEFEDLGVATSGVSLFRTIGGSFGAAIFGSLFVNFLNSRLA
Mb1908|M.bovis_AF2122/97 VPADVADGRVDATAAAIAVPGRPRCRHRIVHAGARSHRAEHVVFRRPRRR
MAV_0138|M.avium_104 --------------EGLIDLEAAAPQRLRGGLTGLFYVVTYIGFGLPLIL
MMAR_0690|M.marinum_M --------------TIMPALADAMESAGGAQRPGTAAAAYNLAFSAGTVA
: :
Mflv_1450|M.gilvum_PYR-GCK AGWAATRWELA-------------------------SVFPWFAGIVSAAC
Mvan_5329|M.vanbaalenii_PYR-1 AGWAATRWDLA-------------------------AVFPWFAGVVAAAC
MSMEG_5167|M.smegmatis_MC2_155 EGLSAQHWGLR-------------------------PAGIAFALAVCVLA
TH_0476|M.thermoresistible__bu EGWAAHHWGLR-------------------------TAGISFAVGVAVLA
MAB_1299c|M.abscessus_ATCC_199 EGLAAQAWGLR-------------------------TAGVTFALAVAALA
Rv1877|M.tuberculosis_H37Rv SALTSGAVPVPAVPSPAVLHQLPQSMAAPIVRAYAESLTQVFLCAVSVTV
MUL_3826|M.ulcerans_Agy99 SALTATEVAS----SPEALHRQSPAVAAPIVHAYAGSLAQVFLCAAPVAA
Mb1908|M.bovis_AF2122/97 -NIGCDLLPGG---RRLVWYRNIRCVVRKLPGPKTRFRADVGRRACPGSA
MAV_0138|M.avium_104 ASVRPG-------------------------------VATAILSGMAVLA
MMAR_0690|M.marinum_M GGPLAGWMADLR-----------------------SFQDAAMLIAVVTLT
Mflv_1450|M.gilvum_PYR-GCK LGAAVAGLRSIRVARAA---------------------------------
Mvan_5329|M.vanbaalenii_PYR-1 MGAAIAGVRSVRLAQ-----------------------------------
MSMEG_5167|M.smegmatis_MC2_155 VICLVAILVEESRQRRAESAELSAST------------------------
TH_0476|M.thermoresistible__bu IGCLIAILIQESRDPQAI--------------------------------
MAB_1299c|M.abscessus_ATCC_199 ALCLIAVIWQERPSRTEALAPVSTG-------------------------
Rv1877|M.tuberculosis_H37Rv VGFILALLLREVPLTDIHDDADDLGDGFGVPRAESPEDVLEIAVRRMLPN
MUL_3826|M.ulcerans_Agy99 IGFVLALFMREVPLRDIHNRDHDAR----VPSGD----------------
Mb1908|M.bovis_AF2122/97 ISGCLASAAPEHGRPDRAGICRVAHPG--VPLRG----------------
MAV_0138|M.avium_104 VTAAVGRAARLRRDDHRQN-------------------------------
MMAR_0690|M.marinum_M CGVLAALVHAAVVRRAVSDA------------------------------
.
Mflv_1450|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5329|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5167|M.smegmatis_MC2_155 --------------------------------------------------
TH_0476|M.thermoresistible__bu --------------------------------------------------
MAB_1299c|M.abscessus_ATCC_199 --------------------------------------------------
Rv1877|M.tuberculosis_H37Rv GVRLRDIATQPGCGLGVAELWALLRIYQYQRLFEAVRLTDIGRHLHVPYQ
MUL_3826|M.ulcerans_Agy99 -----------GSGQ-----------------------------------
Mb1908|M.bovis_AF2122/97 --------------------------------------------------
MAV_0138|M.avium_104 --------------------------------------------------
MMAR_0690|M.marinum_M --------------------------------------------------
Mflv_1450|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5329|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_5167|M.smegmatis_MC2_155 --------------------------------------------------
TH_0476|M.thermoresistible__bu --------------------------------------------------
MAB_1299c|M.abscessus_ATCC_199 --------------------------------------------------
Rv1877|M.tuberculosis_H37Rv VFEPVFDRLVQTGYAARDGDILTLTPSGHRQVDSLAVLIRQWLLDHLAVA
MUL_3826|M.ulcerans_Agy99 ----------QN---KLEGDLDTGQSVG---------------------A
Mb1908|M.bovis_AF2122/97 --------------LGHGGRFHPGAVAAR---------------------
MAV_0138|M.avium_104 --------------------------------------------------
MMAR_0690|M.marinum_M --------------------------------------------------
Mflv_1450|M.gilvum_PYR-GCK ---------------------------------------------
Mvan_5329|M.vanbaalenii_PYR-1 ---------------------------------------------
MSMEG_5167|M.smegmatis_MC2_155 ---------------------------------------------
TH_0476|M.thermoresistible__bu ---------------------------------------------
MAB_1299c|M.abscessus_ATCC_199 ---------------------------------------------
Rv1877|M.tuberculosis_H37Rv PGLKRQPDHQFEAALQHVTDAVLVQRDWYEDLGDLSESRQLAATT
MUL_3826|M.ulcerans_Agy99 PGK--EPDFGVDPAMGPVR--------------------------
Mb1908|M.bovis_AF2122/97 -----------GTAHRHPR--------------------------
MAV_0138|M.avium_104 ---------------------------------------------
MMAR_0690|M.marinum_M ---------------------------------------------