For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
CYAYFVRSRYDQLDDLLTRLHLARQRPQWRRRVLGPDSPVSGVSTIRVLRAVEQFDESGGASVRDVADFL AVEQSTASRLVAPAVAAGLLTKSSAADDQRRRTLVLTEVGRKELAEIAERRRELVAETVADLPDSDVDTL VELLGRITDRFERAAR*
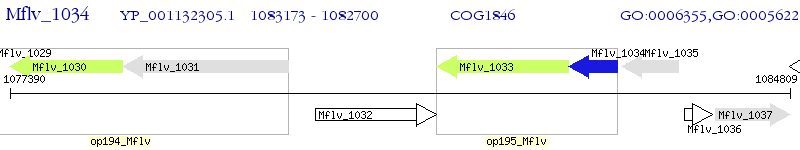
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1034 | - | - | 100% (157) | MarR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_00550 | - | 4e-05 | 27.00% (100) | putative marR-family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_1138c | - | 8e-06 | 29.35% (92) | MarR family transcriptional regulator |
| M. marinum M | MMAR_2213 | - | 1e-06 | 26.92% (104) | transcriptional regulatory protein |
| M. avium 104 | MAV_5180 | - | 9e-07 | 34.04% (94) | MarR-family protein transcriptional regulator, putative |
| M. smegmatis MC2 155 | MSMEG_1330 | - | 3e-43 | 60.96% (146) | MarR-family protein transcriptional regulator |
| M. thermoresistible (build 8) | TH_1752 | - | 2e-47 | 62.18% (156) | MarR-family protein transcriptional regulator |
| M. ulcerans Agy99 | MUL_1800 | - | 1e-06 | 26.92% (104) | transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_1330|M.smegmatis_MC2_155 --------------------------------------------------
TH_1752|M.thermoresistible__bu --------------------------------------------------
Mflv_1034|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_2213|M.marinum_M --------------------------------------------------
MUL_1800|M.ulcerans_Agy99 MARTAIANKAISEYLRPRGSLGSGTEANTSRNSDPMSSLRSPALQWSNTS
MLBr_00550|M.leprae_Br4923 --------------------------------------------------
MAV_5180|M.avium_104 --------------------------------------------------
MAB_1138c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_1330|M.smegmatis_MC2_155 --------------------------------------------------
TH_1752|M.thermoresistible__bu --------------------------------------------------
Mflv_1034|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_2213|M.marinum_M --------------------------------------------------
MUL_1800|M.ulcerans_Agy99 GTDDTGNTETVGTGNSRPTTNSNQLDRSPDSPCLHCGPNSAAPKIINPAH
MLBr_00550|M.leprae_Br4923 --------------------------------------------------
MAV_5180|M.avium_104 --------------------------------------------------
MAB_1138c|M.abscessus_ATCC_199 --------------------------------------------------
MSMEG_1330|M.smegmatis_MC2_155 ----------------------MPRYNELDELLVRVHTVRQRPGWRRRLI
TH_1752|M.thermoresistible__bu --------------MCYAYLVASSRYDDLDELLTRILITRQRPDWRRRLL
Mflv_1034|M.gilvum_PYR-GCK --------------MCYAYFVRS-RYDQLDDLLTRLHLARQRPQWRRRVL
MMAR_2213|M.marinum_M ----------------MSTENATAAEESLDMITDALLTASR--LLVGISA
MUL_1800|M.ulcerans_Agy99 KPANQQRRSPGGRGPLMSTENATAAEESLDMITDALPTASR--LLVGISA
MLBr_00550|M.leprae_Br4923 ----------------MVTDDATVADESLDDITDALLTASR--LLVAVSA
MAV_5180|M.avium_104 -------------------------------------MVGR---FRRQLR
MAB_1138c|M.abscessus_ATCC_199 ---------------------------MTDALVDGLRSFNRTVTQRIGVL
:
MSMEG_1330|M.smegmatis_MC2_155 DDTGSVPSLSTLRVLR-AVEQKEKTGQGASIGEVAEYMAVEHSTASRSVA
TH_1752|M.thermoresistible__bu DSAGPVTSMSSLRALR-AVELRERAGQAASIGDVAEFLAVDHSTASRTVS
Mflv_1034|M.gilvum_PYR-GCK GPDSPVSGVSTIRVLR-AVEQFDESG-GASVRDVADFLAVEQSTASRLVA
MMAR_2213|M.marinum_M HSIAQVDENITIPQFR-TLVILSNRG-PVNLATLANLLGVQPSATGRMVD
MUL_1800|M.ulcerans_Agy99 HSIAQVDENITIPQFR-TLVILSNRG-PVNLATLANLLGVQPSATGRMVD
MLBr_00550|M.leprae_Br4923 HSIGQVDETITFPQFR-TLVILSNRG-PVNLATLAGLLGVQPSATGRMVD
MAV_5180|M.avium_104 RSAGRAFDPARLSESQSELLWLVGRRPGISVSAAAAELGLVPNTASTLVT
MAB_1138c|M.abscessus_ATCC_199 ES-EYLARDRPLGQSR---VLWEIGDSGCDIRELRARLGLDSGYLSRLLR
: : .: :.: . . :
MSMEG_1330|M.smegmatis_MC2_155 NVVAAGLLTKTLSPDDQRRCVLVLTDVGRKALSEVTGRRRQMVAETVADW
TH_1752|M.thermoresistible__bu AIVVAGLLTKQPAADDQRRTVLTLTDVGRKTLAQVTERRREMVTEVVADW
Mflv_1034|M.gilvum_PYR-GCK PAVAAGLLTKSSAADDQRRRTLVLTEVGRKELAEIAERRRELVAETVADL
MMAR_2213|M.marinum_M RLVSAGMIDRQPHPTSRRELLAVLTKRGRDLVRRVTAHRRDEIARIVEQM
MUL_1800|M.ulcerans_Agy99 RLVSAGMIDRQPHPTSRRELLAVLTKRGRDLVRRVTAHRRDEIARIVEQM
MLBr_00550|M.leprae_Br4923 RLVAARLIDRLPHPTSRRELLVALTKRGREVVRRVTAQRRVEIARIVEKM
MAV_5180|M.avium_104 KLVSGGFLLRAAADTDRRVCQLRLTESAQQIVDESRAARRALLSEVVDEL
MAB_1138c|M.abscessus_ATCC_199 ALEGAGLIEVTAADGDTRVRTARLTAAGRAELAELDRRSDDLAHSILEPL
. :: . * ** .: : . :
MSMEG_1330|M.smegmatis_MC2_155 PAEDVDTLVGLLERLVTDFERGVCS-------------------------
TH_1752|M.thermoresistible__bu PDSDVDTLVSLLQRLATDLERGVSQ-------------------------
Mflv_1034|M.gilvum_PYR-GCK PDSDVDTLVELLGRITDRFERAAR--------------------------
MMAR_2213|M.marinum_M PPAERQGLVRALTAFTAAGGEPDANVEMDL--------------------
MUL_1800|M.ulcerans_Agy99 PPAERQGLVRALTAFTAAGGEPDANVEMDL--------------------
MLBr_00550|M.leprae_Br4923 PPSERLGLVRALTAFSTAGGEPDVRLDIISI-------------------
MAV_5180|M.avium_104 NDDQIEALTRGLEVLDMMTQKLRERRP-----------------------
MAB_1138c|M.abscessus_ATCC_199 SVNQRQRLADAMAEVERLFVASQVQIAVMDTRTPEARYCIRAYFEEISAR
: *. : .
MSMEG_1330|M.smegmatis_MC2_155 --------------------------------------------------
TH_1752|M.thermoresistible__bu --------------------------------------------------
Mflv_1034|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_2213|M.marinum_M --------------------------------------------------
MUL_1800|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00550|M.leprae_Br4923 --------------------------------------------------
MAV_5180|M.avium_104 --------------------------------------------------
MAB_1138c|M.abscessus_ATCC_199 FEDGFDPALTLPAGDAELCAPAGLFLVAALHEEPVGCAGLKLPPHTDTAE
MSMEG_1330|M.smegmatis_MC2_155 --------------------------------------------------
TH_1752|M.thermoresistible__bu --------------------------------------------------
Mflv_1034|M.gilvum_PYR-GCK --------------------------------------------------
MMAR_2213|M.marinum_M --------------------------------------------------
MUL_1800|M.ulcerans_Agy99 --------------------------------------------------
MLBr_00550|M.leprae_Br4923 --------------------------------------------------
MAV_5180|M.avium_104 --------------------------------------------------
MAB_1138c|M.abscessus_ATCC_199 IKRVWIAPHVRGLGLGKRILAELERLAVDHGAHTIRLDTNRALTEAIAMY
MSMEG_1330|M.smegmatis_MC2_155 --------------------------
TH_1752|M.thermoresistible__bu --------------------------
Mflv_1034|M.gilvum_PYR-GCK --------------------------
MMAR_2213|M.marinum_M --------------------------
MUL_1800|M.ulcerans_Agy99 --------------------------
MLBr_00550|M.leprae_Br4923 --------------------------
MAV_5180|M.avium_104 --------------------------
MAB_1138c|M.abscessus_ATCC_199 RSAGYDEVAAFNQEKYAHFWFQKQLS