For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
PRYNELDELLVRVHTVRQRPGWRRRLIDDTGSVPSLSTLRVLRAVEQKEKTGQGASIGEVAEYMAVEHST ASRSVANVVAAGLLTKTLSPDDQRRCVLVLTDVGRKALSEVTGRRRQMVAETVADWPAEDVDTLVGLLER LVTDFERGVCS*
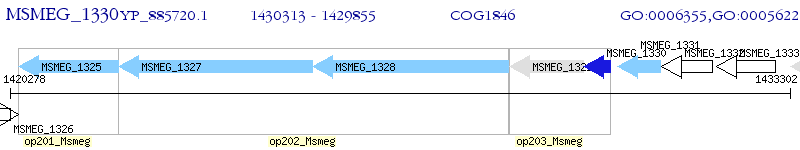
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_1330 | - | - | 100% (152) | MarR-family protein transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_5566 | - | 1e-06 | 28.97% (107) | transcriptional regulator |
| M. smegmatis MC2 155 | MSMEG_4471 | - | 2e-05 | 28.00% (100) | MarR-family protein transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1034 | - | 2e-43 | 60.96% (146) | MarR family transcriptional regulator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | MLBr_00550 | - | 2e-06 | 27.97% (118) | putative marR-family transcriptional regulator |
| M. abscessus ATCC 19977 | MAB_1021 | - | 2e-07 | 30.70% (114) | MarR family transcriptional regulator |
| M. marinum M | MMAR_0125 | - | 5e-07 | 31.13% (106) | MarR family transcriptional regulator |
| M. avium 104 | MAV_3374 | - | 8e-08 | 30.09% (113) | transcriptional regulator, MarR family protein |
| M. thermoresistible (build 8) | TH_1752 | - | 9e-53 | 66.89% (148) | MarR-family protein transcriptional regulator |
| M. ulcerans Agy99 | MUL_0657 | - | 4e-07 | 28.57% (91) | transcriptional regulator |
| M. vanbaalenii PYR-1 | Mvan_5020 | - | 3e-05 | 24.73% (93) | MarR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_1034|M.gilvum_PYR-GCK ------------------------------------MCYAYFVR-SRYDQ
TH_1752|M.thermoresistible__bu ------------------------------------MCYAYLVASSRYDD
MSMEG_1330|M.smegmatis_MC2_155 --------------------------------------------MPRYNE
MLBr_00550|M.leprae_Br4923 ---------------------------------------MVTDDATVADE
MAV_3374|M.avium_104 ----------------------------------------MAADHTTVDE
MAB_1021|M.abscessus_ATCC_1997 ----------------------------------------MTVDDVDLDE
MMAR_0125|M.marinum_M ------------------------------------MVVAMSGGGSPGKQ
MUL_0657|M.ulcerans_Agy99 MAGCRGPAIYTLLTMSSSCRYCRVHAGLHYRRVRAGQTGANVASARKLTA
Mvan_5020|M.vanbaalenii_PYR-1 -----------------------------------------MSGNPLADD
Mflv_1034|M.gilvum_PYR-GCK LDDLLTRLHLARQRPQWRRRVLGPDSPVSGVSTIRVLRAVEQFDESG-GA
TH_1752|M.thermoresistible__bu LDELLTRILITRQRPDWRRRLLDSAGPVTSMSSLRALRAVELRERAGQAA
MSMEG_1330|M.smegmatis_MC2_155 LDELLVRVHTVRQRPGWRRRLIDDTGSVPSLSTLRVLRAVEQKEKTGQGA
MLBr_00550|M.leprae_Br4923 SLDDITDALLTASR-LLVAVSAHSIGQVDETITFPQFRTLVILSNRG-PV
MAV_3374|M.avium_104 SLDAITDALLTASR-LLMAISARSVGQVDETITIPQFRTLVILSNRG-PV
MAB_1021|M.abscessus_ATCC_1997 SLDALADSLLVASR-LLLAISARSIAQVDETITFPQFRTLVILFNEG-PI
MMAR_0125|M.marinum_M LADDLRDALVQTSF---ALMAVLTEVAAEHDLSLTQLRVLGIMRDRE--P
MUL_0657|M.ulcerans_Agy99 VAAISATLAQIVRLSISRSAFAGQASAANTELSQPSYVLLRVLIDEG-PL
Mvan_5020|M.vanbaalenii_PYR-1 VWRDLAALVLDHRD-GWKRAVVEQSG-----LPFSRIRILKRLSKTP--M
. :
Mflv_1034|M.gilvum_PYR-GCK SVRDVADFLAVEQSTASRLVAPAVAAGLLTKSSAADDQRRRTLVLTEVGR
TH_1752|M.thermoresistible__bu SIGDVAEFLAVDHSTASRTVSAIVVAGLLTKQPAADDQRRTVLTLTDVGR
MSMEG_1330|M.smegmatis_MC2_155 SIGEVAEYMAVEHSTASRSVANVVAAGLLTKTLSPDDQRRCVLVLTDVGR
MLBr_00550|M.leprae_Br4923 NLATLAGLLGVQPSATGRMVDRLVAARLIDRLPHPTSRRELLVALTKRGR
MAV_3374|M.avium_104 NLATLAGLLGVQPSATGRMVDRLVAAGLIDRLPHPTSRRELLAALTTRGR
MAB_1021|M.abscessus_ATCC_1997 NLATLAQSLGVQPSTTGRMVDRLVSAGLIDRRPHPRSRRELVAELTAHGE
MMAR_0125|M.marinum_M TMGELASYTGLERSTVSGLIERAAARGLVVKTADENDGRAVRVNLTTGAR
MUL_0657|M.ulcerans_Agy99 PLSRLARLAHMDAGMATRRVRALVEAGLVTRHTDPNDGRVSVIEATPQGR
Mvan_5020|M.vanbaalenii_PYR-1 SVKQLAEAATIDAPAATVAVNDLEQRGLVVRTVDPSNRRCKTVSLTEAGR
: :* :: . : *: : . * * ..
Mflv_1034|M.gilvum_PYR-GCK KELAEIAERRRELVAETVADLPDSDVDTLVELLGRITDRFERAAR-----
TH_1752|M.thermoresistible__bu KTLAQVTERRREMVTEVVADWPDSDVDTLVSLLQRLATDLERGVSQ----
MSMEG_1330|M.smegmatis_MC2_155 KALSEVTGRRRQMVAETVADWPAEDVDTLVGLLERLVTDFERGVCS----
MLBr_00550|M.leprae_Br4923 EVVRRVTAQRRVEIARIVEKMPPSERLGLVRALTAFSTAGGEPDVRLD-I
MAV_3374|M.avium_104 DVVHQVTAHRRAEIAAIVAKMPPAERHGLVRALTAFTAAGGEPDVHLDAE
MAB_1021|M.abscessus_ATCC_1997 GVVREVMAQRRAEIAEIAARMPARERRGLVRALDAFTAAGGESASPVDV-
MMAR_0125|M.marinum_M RLERETTKAIEQRIAPMLGRLSTREQQRLAALLSKTLER-----------
MUL_0657|M.ulcerans_Agy99 RAAGALHEVRRDHLARALSGWSAAELHEFNRLLSKFLTDIKKTPIVDTAT
Mvan_5020|M.vanbaalenii_PYR-1 DTLRAIDAVH-DPAPDMLAALGDDDLAALRVILDKLDAG-----------
. : : *
Mflv_1034|M.gilvum_PYR-GCK ---
TH_1752|M.thermoresistible__bu ---
MSMEG_1330|M.smegmatis_MC2_155 ---
MLBr_00550|M.leprae_Br4923 ISI
MAV_3374|M.avium_104 IDL
MAB_1021|M.abscessus_ATCC_1997 ---
MMAR_0125|M.marinum_M ---
MUL_0657|M.ulcerans_Agy99 R--
Mvan_5020|M.vanbaalenii_PYR-1 ---