For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VVGRFRRQLRRSAGRAFDPARLSESQSELLWLVGRRPGISVSAAAAELGLVPNTASTLVTKLVSGGFLLR AAADTDRRVCQLRLTESAQQIVDESRAARRALLSEVVDELNDDQIEALTRGLEVLDMMTQKLRERRP
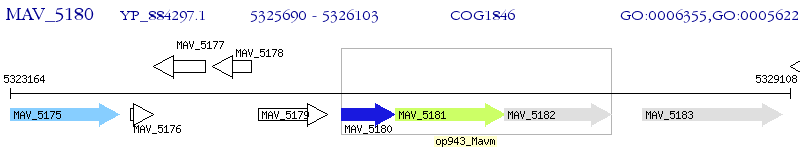
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. avium 104 | MAV_5180 | - | - | 100% (137) | MarR-family protein transcriptional regulator, putative |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. gilvum PYR-GCK | Mflv_1034 | - | 8e-07 | 34.04% (94) | MarR family transcriptional regulator |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2648c | - | 5e-07 | 27.91% (129) | MarR family transcriptional regulator |
| M. marinum M | MMAR_2213 | - | 1e-05 | 26.80% (97) | transcriptional regulatory protein |
| M. smegmatis MC2 155 | MSMEG_6508 | - | 5e-40 | 62.22% (135) | MarR-family protein transcriptional regulator |
| M. thermoresistible (build 8) | TH_1752 | - | 3e-05 | 28.69% (122) | MarR-family protein transcriptional regulator |
| M. ulcerans Agy99 | MUL_1800 | - | 9e-06 | 26.80% (97) | transcriptional regulatory protein |
| M. vanbaalenii PYR-1 | Mvan_4913 | - | 1e-06 | 30.16% (126) | MarR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
MAV_5180|M.avium_104 --------------------------------------------------
MSMEG_6508|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1034|M.gilvum_PYR-GCK --------------------------------------------------
TH_1752|M.thermoresistible__bu --------------------------------------------------
MMAR_2213|M.marinum_M --------------------------------------------------
MUL_1800|M.ulcerans_Agy99 MARTAIANKAISEYLRPRGSLGSGTEANTSRNSDPMSSLRSPALQWSNTS
Mvan_4913|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_2648c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_5180|M.avium_104 --------------------------------------------------
MSMEG_6508|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_1034|M.gilvum_PYR-GCK --------------------------------------------------
TH_1752|M.thermoresistible__bu --------------------------------------------------
MMAR_2213|M.marinum_M --------------------------------------------------
MUL_1800|M.ulcerans_Agy99 GTDDTGNTETVGTGNSRPTTNSNQLDRSPDSPCLHCGPNSAAPKIINPAH
Mvan_4913|M.vanbaalenii_PYR-1 --------------------------------------------------
MAB_2648c|M.abscessus_ATCC_199 --------------------------------------------------
MAV_5180|M.avium_104 -----------------------------------MVGRFRRQLRRSAGR
MSMEG_6508|M.smegmatis_MC2_155 ---------------MTETQRPLDTEVIGDLAS--VIGRFRRQLRRATGG
Mflv_1034|M.gilvum_PYR-GCK -------------MCYAYFVR-SRYDQLDDLLTRLHLARQRPQWRRRVLG
TH_1752|M.thermoresistible__bu -------------MCYAYLVASSRYDDLDELLTRILITRQRPDWRRRLLD
MMAR_2213|M.marinum_M ----------------MSTENATAAEESLDMITDALLTASR-LLVGISAH
MUL_1800|M.ulcerans_Agy99 KPANQQRRSPGGRGPLMSTENATAAEESLDMITDALPTASR-LLVGISAH
Mvan_4913|M.vanbaalenii_PYR-1 ----------------MGGRAASGASAQRAGRLADAFGRAGKSVVRAFDD
MAB_2648c|M.abscessus_ATCC_199 ------MAVHESSITTRDWTLNRPELDTSPMEVIGALKRATGMLARLLEP
MAV_5180|M.avium_104 AFDPARLSESQSELLWLVGR-RPG--ISVSAAAAELGLVPNTASTLVTKL
MSMEG_6508|M.smegmatis_MC2_155 GFDAAGLTQSQGELLRLVGR-KPG--ISVRESADELSLVPNTASTLVSRL
Mflv_1034|M.gilvum_PYR-GCK PDSPVSGVSTIRVLRAVEQFDESG-GASVRDVADFLAVEQSTASRLVAPA
TH_1752|M.thermoresistible__bu SAGPVTSMSSLRALRAVELRERAGQAASIGDVAEFLAVDHSTASRTVSAI
MMAR_2213|M.marinum_M SIAQVDENITIPQFRTLVILSNRG-PVNLATLANLLGVQPSATGRMVDRL
MUL_1800|M.ulcerans_Agy99 SIAQVDENITIPQFRTLVILSNRG-PVNLATLANLLGVQPSATGRMVDRL
Mvan_4913|M.vanbaalenii_PYR-1 RLGEHGVSTPRSKLLAEVARMQPV---RLADLAREVGISQGTASTLVEAL
MAB_2648c|M.abscessus_ATCC_199 LYATSAIAEPEHDVLVVLRHRKGP--VIARHVAEELGVSQAWVSRMLRKL
. . . * :.: .. :
MAV_5180|M.avium_104 VSGGFLLRAAADTDRRVCQLRLTESAQQIVDESRAARRALLSEVVDELND
MSMEG_6508|M.smegmatis_MC2_155 VADGLLVKTADESDRRVGRLWLTPRAQRIADESRAARRAALSDVLAKLDD
Mflv_1034|M.gilvum_PYR-GCK VAAGLLTKSSAADDQRRRTLVLTEVGRKELAEIAERRRELVAETVADLPD
TH_1752|M.thermoresistible__bu VVAGLLTKQPAADDQRRTVLTLTDVGRKTLAQVTERRREMVTEVVADWPD
MMAR_2213|M.marinum_M VSAGMIDRQPHPTSRRELLAVLTKRGRDLVRRVTAHRRDEIARIVEQMPP
MUL_1800|M.ulcerans_Agy99 VSAGMIDRQPHPTSRRELLAVLTKRGRDLVRRVTAHRRDEIARIVEQMPP
Mvan_4913|M.vanbaalenii_PYR-1 VRDGLVARGVDDNDRRAIRLTTTPEGETQARNWLRDYVVVAGEIFSCLTA
MAB_2648c|M.abscessus_ATCC_199 EERGYVRREANANDRRAAIITMTDSGRSVVDELFPERLRIEAEALAGLGD
* : : .:* * .. . .
MAV_5180|M.avium_104 DQIEALTRGLEVLDMMTQKLRERRP---
MSMEG_6508|M.smegmatis_MC2_155 DQIEHLAKGLDVLAELTRLLQERDS---
Mflv_1034|M.gilvum_PYR-GCK SDVDTLVE---LLGRITDRFERAAR---
TH_1752|M.thermoresistible__bu SDVDTLVS---LLQRLATDLERGVSQ--
MMAR_2213|M.marinum_M AERQGLVRALTAFTAAGGEPDANVEMDL
MUL_1800|M.ulcerans_Agy99 AERQGLVRALTAFTAAGGEPDANVEMDL
Mvan_4913|M.vanbaalenii_PYR-1 EEQRQLTR---LLDRIAESLDSPA----
MAB_2648c|M.abscessus_ATCC_199 ERAAVVAG----LERLVATLKAYEG---
:. :