For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SSSLGDVLASMQLRSVGDGLFAGAQLPAPANHILGGHISAQALLAASLTAPGRAPHSVHTYFLRPGDSRH AVTFEVVKLQEGRTFSARRVTARQGDEILLEALSSFKLPTEAPGGVTYQPPCPDVPAPESLPVVAPHSAE SHSAEGTWASLRWFERRVVDAATGPPAPARIWLRPDGEVPDDPVLTASLVAYLSAVTLTEPAYAARSGLV MAAQRDHSVWFHAAADLSDWMLYDMASPSSADTLALARGTLFNRSGELVCTVRQEMYFPPPRG*
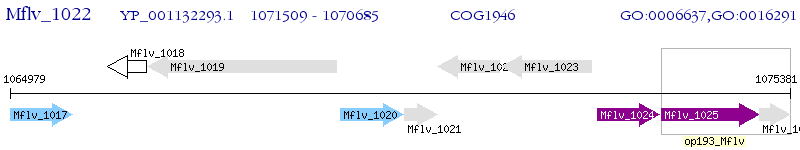
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1022 | - | - | 100% (274) | acyl-CoA thioesterase |
| M. gilvum PYR-GCK | Mflv_4093 | - | 4e-30 | 34.72% (265) | acyl-CoA thioesterase |
| M. gilvum PYR-GCK | Mflv_3831 | - | 9e-30 | 32.02% (253) | acyl-CoA thioesterase II |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2637c | tesB2 | 2e-33 | 34.73% (239) | acyl-CoA thioesterase II |
| M. tuberculosis H37Rv | Rv2605c | tesB2 | 5e-34 | 35.15% (239) | acyl-CoA thioesterase II |
| M. leprae Br4923 | MLBr_01278 | tesB | 2e-27 | 29.43% (299) | acyl CoA thioesterase II |
| M. abscessus ATCC 19977 | MAB_2891c | - | 7e-34 | 32.33% (266) | acyl-CoA thioesterase II TesB2 |
| M. marinum M | MMAR_2095 | tesB2 | 2e-31 | 33.87% (248) | acyl-CoA thioesterase II TesB2 |
| M. avium 104 | MAV_2695 | - | 4e-80 | 63.60% (239) | acyl-CoA thioesterase family protein |
| M. smegmatis MC2 155 | MSMEG_3812 | - | 4e-62 | 45.29% (276) | acyl-CoA thioesterase |
| M. thermoresistible (build 8) | TH_3497 | - | 2e-56 | 42.86% (280) | PUTATIVE acyl-CoA thioesterase |
| M. ulcerans Agy99 | MUL_3249 | tesB2 | 2e-31 | 33.87% (248) | acyl-CoA thioesterase II TesB2 |
| M. vanbaalenii PYR-1 | Mvan_5814 | - | 1e-124 | 81.02% (274) | acyl-CoA thioesterase |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_2095|M.marinum_M --------------------------------------------------
MUL_3249|M.ulcerans_Agy99 --------------------------------------------------
Mb2637c|M.bovis_AF2122/97 --------------------------------------------------
Rv2605c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_2891c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_01278|M.leprae_Br4923 --------------------------------------------------
Mflv_1022|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5814|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_2695|M.avium_104 --------------------------------------------------
MSMEG_3812|M.smegmatis_MC2_155 --------------------------------------------------
TH_3497|M.thermoresistible__bu LIRNVRPYGEGEPVDVLVADGQIAEIGEKLPIPGDGELGDVIDADGQILL
MMAR_2095|M.marinum_M --------------------------------------------------
MUL_3249|M.ulcerans_Agy99 --------------------------------------------------
Mb2637c|M.bovis_AF2122/97 --------------------------------------------------
Rv2605c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_2891c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_01278|M.leprae_Br4923 --------------------------------------------------
Mflv_1022|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5814|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_2695|M.avium_104 --------------------------------------------------
MSMEG_3812|M.smegmatis_MC2_155 --------------------------------------------------
TH_3497|M.thermoresistible__bu PGFVDLHTHLREPGREYAEDIETGSAAAALGGYTAVFAMANTDPVADSPV
MMAR_2095|M.marinum_M --------------------------------------------------
MUL_3249|M.ulcerans_Agy99 --------------------------------------------------
Mb2637c|M.bovis_AF2122/97 --------------------------------------------------
Rv2605c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_2891c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_01278|M.leprae_Br4923 --------------------------------------------------
Mflv_1022|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5814|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_2695|M.avium_104 --------------------------------------------------
MSMEG_3812|M.smegmatis_MC2_155 --------------------------------------------------
TH_3497|M.thermoresistible__bu VTDHVWRRGQQVGLVDVHPVGAVTVGLEGRQLTEMGLMAAGAARVRMFSD
MMAR_2095|M.marinum_M --------------------------------------------------
MUL_3249|M.ulcerans_Agy99 --------------------------------------------------
Mb2637c|M.bovis_AF2122/97 --------------------------------------------------
Rv2605c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_2891c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_01278|M.leprae_Br4923 --------------------------------------------------
Mflv_1022|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5814|M.vanbaalenii_PYR-1 --------------------------------------------------
MAV_2695|M.avium_104 --------------------------------------------------
MSMEG_3812|M.smegmatis_MC2_155 --------------------------------------------------
TH_3497|M.thermoresistible__bu DGLCVHDPLVMRRALEYARGLGVLIAQHAEEPRLTVGAVAHXXXWPACPA
MMAR_2095|M.marinum_M --------------------------------------------------
MUL_3249|M.ulcerans_Agy99 --------------------------------------------------
Mb2637c|M.bovis_AF2122/97 --------------------------------------------------
Rv2605c|M.tuberculosis_H37Rv --------------------------------------------------
MAB_2891c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_01278|M.leprae_Br4923 --------------------------------------------------
Mflv_1022|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_5814|M.vanbaalenii_PYR-1 ------------------------------------MRSPTMKSSRTTIK
MAV_2695|M.avium_104 --------------------------------------------------
MSMEG_3812|M.smegmatis_MC2_155 ------------------------------------MPLP----------
TH_3497|M.thermoresistible__bu RCRPGYRSPPTWCASAPPGRPNAVRRNRSSPRRIGPTPPPGTSRPATDVG
MMAR_2095|M.marinum_M -----------------------------MAIEEILDLEQLEVNIYRGSV
MUL_3249|M.ulcerans_Agy99 -----------------------------MAIEEILDLEQLEVNIYRGSV
Mb2637c|M.bovis_AF2122/97 -----------------------------MSIEEILDLEQLEVNIYRGSV
Rv2605c|M.tuberculosis_H37Rv -----------------------------MSIEEILDLEQLEVNIYRGSV
MAB_2891c|M.abscessus_ATCC_199 ----------------------------MADIEEILTLEQLEKDIFRGNV
MLBr_01278|M.leprae_Br4923 -------------------MKRLWEKKTMSDFEELLAVLDLNRVADDLFI
Mflv_1022|M.gilvum_PYR-GCK ---------------------------MSSSLGDVLASMQLRSVGDGLFA
Mvan_5814|M.vanbaalenii_PYR-1 RRGTSRDEEPDHG--------VRQDTCVSSSLGDVLASMQLDQAGPAAFV
MAV_2695|M.avium_104 ---------------------------MIRSPASAVRSASSTSS-KADWS
MSMEG_3812|M.smegmatis_MC2_155 ------------------------------QLADVLATLDITDCGHDMFV
TH_3497|M.thermoresistible__bu RLGSADRARRPGPRCRGAWWLGHDAPVTELTLYDVLTTLDVAQVDDDLFE
. : .
MMAR_2095|M.marinum_M FSPESGFLQRTFGGHVAGQSLVSAVRTVDPRYQVHSLHG-YFLRSGDAQE
MUL_3249|M.ulcerans_Agy99 FSPESGFLQRTFGGHVAGQSLVSAVRTVDPRYQVHSLHG-YFLRSGDAQE
Mb2637c|M.bovis_AF2122/97 FSPESGFLQRTFGGHVAGQSLVSAVRTVDPRYMVHSLHG-YFLRPGDAKE
Rv2605c|M.tuberculosis_H37Rv FSPESGFLQRTFGGHVAGQSLVSAVRTVDPRYMVHSLHG-YFLRPGDAKE
MAB_2891c|M.abscessus_ATCC_199 TPSN---LRRTFGGQVAGQSLVSAVRTVEPEYLVHSLHG-YFLRPGNPNE
MLBr_01278|M.leprae_Br4923 GSHPSKNPLRTFGGQLVAQSFVASSRTLIRDDLPPSAFSVHFINGGDTAK
Mflv_1022|M.gilvum_PYR-GCK GAQLPAPANHILGGHISAQALLAASLTAPGR--APHSVHTYFLRPGDSRH
Mvan_5814|M.vanbaalenii_PYR-1 GRQLPAPANHILGGHISAQALLAASLTAPGR--TPHSVHTYFLRPGDSRL
MAV_2695|M.avium_104 RAIAMFLSVVILGRFTTETHAMALLRQGPTR--RHHLHHS----PG--RS
MSMEG_3812|M.smegmatis_MC2_155 ATQIDNEAHHIIGGHISAQALMAASRTAAGR--VPNSLHVYLLRAGDARY
TH_3497|M.thermoresistible__bu ATQLDNPAHHIVGGHIAGQAVVAASRTAPQR--TAHSAHVYFLRAGDARY
.* :: *
MMAR_2095|M.marinum_M PTVFLVERTRDGGSFVTRRVNAVQHGEVIFSMGASFQTA---QNGISHQD
MUL_3249|M.ulcerans_Agy99 PTVFLVERTRDGGSFVTRRVNAVQHGEVIFSMGASFQTA---QNGISHQD
Mb2637c|M.bovis_AF2122/97 RTVFLVERIRDGGSLCTRRVNAVQHGETIFSMAASFQTE---QEGITHQD
Rv2605c|M.tuberculosis_H37Rv RTVFLVERIRDGGSFCTRRVNAVQHGETIFSMAASFQTE---QEGITHQD
MAB_2891c|M.abscessus_ATCC_199 PTVFLVDRVRDGRSFVTRRVTAIQDGQAIFSMSASFQTL---DTGIEHQD
MLBr_01278|M.leprae_Br4923 EIEFHVVRLRDERRFANRRVDAIQDGMLLSSAMASYMSG---GCGLEHAV
Mflv_1022|M.gilvum_PYR-GCK AVTFEVVKLQEGRTFSARRVTARQGDEILLEALSSFKLPTEAPGGVTYQP
Mvan_5814|M.vanbaalenii_PYR-1 TVTFEVVNLQEGRTFSARRVTARQGDEILLEAMSSFKSSTPSARPVTYQP
MAV_2695|M.avium_104 PVDFEVVDLQEGRTFSARRVTARQDDKILMEAMSSFKVVNSAAAQVVYQP
MSMEG_3812|M.smegmatis_MC2_155 PVEMQVMNMRDGGMLSTRRVIARQGDEVLLEALVSLSDT---ADTVDYHQ
TH_3497|M.thermoresistible__bu PVQMRVDRLRDGGSLSTXRVTATQDGQVLLEALASFTVR---IDSVDTHH
: * :: : ** * * . : . * :
MMAR_2095|M.marinum_M AMPAAPPPDDLPGLRSVRVFDDAGFRQFE------EWDVRIVPRDLLAPL
MUL_3249|M.ulcerans_Agy99 AMPAAPPPDDLPGLRSVRVFDDAGFRQFE------EWDVRIVPRDLLAPL
Mb2637c|M.bovis_AF2122/97 VMPAAPPPDGLPGLNSIKVFDDAGFRQFD------EWDVCIVPRERLRLL
Rv2605c|M.tuberculosis_H37Rv VMPAAPPPDGLPGLNSIKVFDDAGFRQFD------EWDVCIVPRERLRLL
MAB_2891c|M.abscessus_ATCC_199 LMPPVPDPEDLPDAQEQDSFNRDLFRQFA------EWDIRIVPDELMDRN
MLBr_01278|M.leprae_Br4923 EPPEAAEPHTRPPIGELLRGYEQIVQHFVNALQPVEWRYVNDPSWVMRNK
Mflv_1022|M.gilvum_PYR-GCK PCPDVPAPESLPVVAPHSAES-HSAEGTWASLRWFERRVVDAAT------
Mvan_5814|M.vanbaalenii_PYR-1 RCPEVAPPESLPRVAPHFAESDEVTKGQWASLRWFERRIVDAAA------
MAV_2695|M.avium_104 IMPEAPSPESLPWVAPHLAEADEGSRGRWASLRWFERRTIETET------
MSMEG_3812|M.smegmatis_MC2_155 PMPDVPAPEDLPSVGELLSDYADELDGFWVKPQWIERRYVDAPPRLALDQ
TH_3497|M.thermoresistible__bu PMPDVPDPDTLPPVTEQLVAYADEFGGLWVRPQPIERRYIDPPPRVAYDL
* .. *. * *
MMAR_2095|M.marinum_M PGKASQQQVWFRHRDPLPDDPVLHICALAYMSDLTLLGSAQVTHLAER--
MUL_3249|M.ulcerans_Agy99 PGKASQQQVWFRHRDPLPDDPVLHICALAYMSDLTLLGSAQVTHLAER--
Mb2637c|M.bovis_AF2122/97 PGKASQQQVWLRHRDPLPDDPVLHICALAYMSDLTLLGSAQVNHLDVR--
Rv2605c|M.tuberculosis_H37Rv PGKASQQQVWLRHRDPLPDDPVLHICALAYMSDLTLLGSAQVNHLDVR--
MAB_2891c|M.abscessus_ATCC_199 PRLAAQQRVWFRCRKHLPDDPVLHICALAYMSDLTLLSSSKVPHRDT---
MLBr_01278|M.leprae_Br4923 GQRLAYNRVWVKALGALPNDPTLHTAAMLYSTDTTVLDSVITTHGLSWGF
Mflv_1022|M.gilvum_PYR-GCK -GPPAPARIWLRPDGEVPDDPVLTASLVAYLSAVTLTEPAYAARSG----
Mvan_5814|M.vanbaalenii_PYR-1 -DPPARSRIWWRPDGEVPDDPVLTASLVAYLSAVTLTEPAYAARGG----
MAV_2695|M.avium_104 -VPPARAPMWWRPDGRVPDDPVLTASLVAYMSAVTLTEPAFAARGG----
MSMEG_3812|M.smegmatis_MC2_155 PEPPERTRMWWRPAESIPNDPVLNCCVLTYASGTSLLETTVTMRRTTQ--
TH_3497|M.thermoresistible__bu PEPPARTRMWWRATEPVPDDPALNSALLTYVTGTSTLEAAMVAHRTTP--
:* : :*:**.* . : * : : . . :
MMAR_2095|M.marinum_M EHLQVASLDHAMWFMRGFRADEWLLYDQSSPSAGGGRALTHGKIFTQGGE
MUL_3249|M.ulcerans_Agy99 EHLQVASLDHAMWFMRGFRADEWLLYDQSSPSAGGGRALTHGKIFTQGGE
Mb2637c|M.bovis_AF2122/97 DQLQVASLDHAMWFMRPFRADEWLLYDQSSPSASGGRALTRGEIFTRSGE
Rv2605c|M.tuberculosis_H37Rv DQLQVASLDHAMWFMRPFRADEWLLYDQSSPSASGGRALTRGEIFTRSGE
MAB_2891c|M.abscessus_ATCC_199 -VLQTASLDHALWFLRPFRADEWMLYDETSPSAGFGRALTQGRIFNRDGT
MLBr_01278|M.leprae_Br4923 DRIFAVSANHSVWFHRQVNFDDWVLYSTSSPVAADSRGLGTGHLFDRSGQ
Mflv_1022|M.gilvum_PYR-GCK -LVMAAQRDHSVWFHAAADLSDWMLYDMASPSSADTLALARGTLFNRSGE
Mvan_5814|M.vanbaalenii_PYR-1 -VTVSAQRDHSVWFHAAADLSDWLLYEQSSPSSADTLALASGTMFNRTGE
MAV_2695|M.avium_104 -VGASAQRDHSVWFHGRAVLSDWLFYDRSSPSSAGSLALASGTMFNRTGE
MSMEG_3812|M.smegmatis_MC2_155 ATAFSALIDHTVWFQRPADLSDWVLSDQVSLSGIHGRGLASGTMYNRSGE
TH_3497|M.thermoresistible__bu LHSFNALIDHALWFHRPAHLTDWVLADQFSPSSLGGRGLATATMYNRTGE
. :*::** :*:: . * . .* . :: : *
MMAR_2095|M.marinum_M LVAAVMQEGLTRYPSGYQPGEK------
MUL_3249|M.ulcerans_Agy99 LVAAVMQEGLTRYPSGYQPGEN------
Mb2637c|M.bovis_AF2122/97 MVAAVMQEGLTRHRRGHRSVGQ------
Rv2605c|M.tuberculosis_H37Rv MVAAVMQEGLTRHRRGHRSVGQ------
MAB_2891c|M.abscessus_ATCC_199 MVAAVVQEGLTRIERNPEQASVARGNMA
MLBr_01278|M.leprae_Br4923 LIATVVQEGVLKYFPASHR---------
Mflv_1022|M.gilvum_PYR-GCK LVCTVRQEMYFPPPRG------------
Mvan_5814|M.vanbaalenii_PYR-1 LVCTVRQEMYFPASRH------------
MAV_2695|M.avium_104 LVCTVKQEMYFPPQTA------------
MSMEG_3812|M.smegmatis_MC2_155 LVCSATQEIYFGR-GRI-----------
TH_3497|M.thermoresistible__bu LVCTATQELYFGRPARSGSRR-------
::.:. **