For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTAPPHRRRNAEEDVVGARVSIGDFAVMTSLSRKALRHYHDIGILEPAHIDSHTGYRFYDTGQVDHAHII RRFRSLGMSIPDIKALLSADGAAARTEIITTHLTQMEVQLQHTRDTVSALRELLSPVSVPSHVEVHAKPA LAVWSVGATIEVSEIDDWFGRSLKTLRDAVTAAGSAALGHQPGGLYDRALFLESRGAATLFVPALHSADP PEGVRAEVLPPAEFAVLRHCGGHDGIDRSYGALGRYVNEHLISHQGPIREHYIGATPSTPTTFTATEICW PIFGATPPPQ
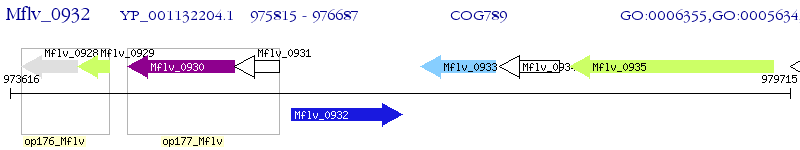
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0932 | - | - | 100% (290) | MerR family transcriptional regulator |
| M. gilvum PYR-GCK | Mflv_3212 | - | 2e-06 | 32.43% (111) | MerR family transcriptional regulator |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3367 | - | 3e-05 | 24.56% (114) | MerR family transcriptional regulator |
| M. tuberculosis H37Rv | Rv3334 | - | 3e-05 | 24.56% (114) | MerR family transcriptional regulator |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_4723c | - | 5e-05 | 27.78% (108) | MerR family transcriptional regulator |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | MAV_1646 | - | 4e-08 | 31.78% (107) | transcriptional regulator, MerR family protein |
| M. smegmatis MC2 155 | MSMEG_3274 | - | 9e-88 | 59.42% (276) | MerR-family protein transcriptional regulator |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_3507 | - | 7e-45 | 36.47% (266) | MerR family transcriptional regulator |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0932|M.gilvum_PYR-GCK MTAPPHRRRNAEEDVVGA-RVSIGDFAVMTSLSRKALRHYHDIGILEPAH
MSMEG_3274|M.smegmatis_MC2_155 ---------------MAG-RVSIGDFAVMTHLSRKALRHYHEMGVLVPAH
Mvan_3507|M.vanbaalenii_PYR-1 MAAICDSAGVKREAVQASPKLTIGEFATVTHLSVRTLRRYHEGGLLEPAW
Mb3367|M.bovis_AF2122/97 --------------------MKISEVAALTNTSTKTLRFYENSGLLPPP-
Rv3334|M.tuberculosis_H37Rv --------------------MKISEVAALTNTSTKTLRFYENSGLLPPP-
MAB_4723c|M.abscessus_ATCC_199 ---------MATDAEGVAVGWSTRELADLAGTSLRTVRHYHDVGLLGEP-
MAV_1646|M.avium_104 ----------------MAEQLLIGDVTALTGIASGRIRHYEKIGLLHAD-
:.: :: : :* *.. *:*
Mflv_0932|M.gilvum_PYR-GCK IDSHTGYRFYDTGQVDHAHIIRRFRSLGMSIPDIKALLS-----ADGAAA
MSMEG_3274|M.smegmatis_MC2_155 IDAHTGYRFYDTCQVDHARIIRRFRELDMPIPDIKALLA-----AGDGAT
Mvan_3507|M.vanbaalenii_PYR-1 VDGYTGYRYYTPEQIPTAQVIHRLRQLDVPLADIESILT-----ADDPQH
Mb3367|M.bovis_AF2122/97 ARTASGYRNYGPEIVDRLRFIHRGQAAGLALQEVRQILA-----IHD---
Rv3334|M.tuberculosis_H37Rv ARTASGYRNYGPEIVDRLRFIHRGQAAGLALQEVRQILA-----IHD---
MAB_4723c|M.abscessus_ATCC_199 ERRPNGYKSYGVAHLVRVLQIKRLTGLGLSLKQIAEMDE-----ADE---
MAV_1646|M.avium_104 -HLSNGYRVFDVEQVLDLLRIDLMRSLGVGINDIQRLLHGGHSTLADLLD
.**: : : * .: : :: :
Mflv_0932|M.gilvum_PYR-GCK RTEIITTHLTQ-MEVQLQHTRDTVSALRELLSPVSVPSHVEVHAKPALAV
MSMEG_3274|M.smegmatis_MC2_155 RNEIIAAHLRR-MEEQLAQTRDAVTELRELLDPVRVPAHIEVRHQPATEV
Mvan_3507|M.vanbaalenii_PYR-1 RAQVISDHLRR-LEAELDRTRAAVVSLHRLLNPEPADLEVQLLSVPARTV
Mb3367|M.bovis_AF2122/97 RGEAPCAHVRQLLSTRIDEVRAQIAELIALEGHLQTLLDHASYGPPTEHD
Rv3334|M.tuberculosis_H37Rv RGEAPCAHVRQLLSTRIDEVRAQIAELIALEGHLQTLLDHASYGPPTEHD
MAB_4723c|M.abscessus_ATCC_199 -------NQQEALRALDAELAATIDRLQGVRDELAQIFSSEIAADLPREF
MAV_1646|M.avium_104 EHRALLIQQRDRLDQLIRALDESCAQSRAAADGTDVSEHLLRQLATTHRD
: : . .
Mflv_0932|M.gilvum_PYR-GCK WSVGATIEVSEIDDWFGRSLKTLRDAVTAAGSAAL---GHQPGGLYDRAL
MSMEG_3274|M.smegmatis_MC2_155 WAVSAEVHSSQIGAWFAASVDTLQRALSDSG-VRI---DGAMGGVYDREL
Mvan_3507|M.vanbaalenii_PYR-1 AGISGHVRQEDSLTWYDSAMAELDAAFPPAE---H---TGPPGGRYDNEL
Mb3367|M.bovis_AF2122/97 HSTVCWILESDLDEPTAIEVSDIHA-------------------------
Rv3334|M.tuberculosis_H37Rv HSTVCWILESDLDEPTAIEVSDIHA-------------------------
MAB_4723c|M.abscessus_ATCC_199 GAAAVDMNFSETDRSLAFVMSRVLG---------------PAGLQAFVGM
MAV_1646|M.avium_104 SIGVIGRLSAPLPAPVAAMYADLFDEWDLPVPALFGQMRLPSAASTLLAQ
:
Mflv_0932|M.gilvum_PYR-GCK FLESRGAATLFVPALHSADPPEGVRAEVLPPAEFAVLRHCGGHDG-----
MSMEG_3274|M.smegmatis_MC2_155 FAGESGVATLYVPVPHTVPVPPTVQAQTVPAQELAVLVHPGSQAG-----
Mvan_3507|M.vanbaalenii_PYR-1 FTHGAGAFAVFRPVRNPKPAG-RIRVLELPQADLAVTVHTGRHDD-----
Mb3367|M.bovis_AF2122/97 --------------------------------------------------
Rv3334|M.tuberculosis_H37Rv --------------------------------------------------
MAB_4723c|M.abscessus_ATCC_199 FLDQQDRALDHELENLPADAGERTRADLAERMSLHAKRLYADHPG-----
MAV_1646|M.avium_104 LAETPGRQVLFDRLRRLAADVIALGESVSAATELAHNWIEQQLADPLPDD
Mflv_0932|M.gilvum_PYR-GCK IDRSYGALGRYVNEHLISHQGPIREHY-IGATPSTPTTFTATEICWP---
MSMEG_3274|M.smegmatis_MC2_155 IDRSYGELGAYVYDHLISHQGPIRERY-VGAPASNIVAFTATEIGWP---
Mvan_3507|M.vanbaalenii_PYR-1 IDVTYGRLGSWVVTHALAVDGPIHETYSVGPRDTTDPAEWRTEIGWP---
Mb3367|M.bovis_AF2122/97 --------------------------------------------------
Rv3334|M.tuberculosis_H37Rv --------------------------------------------------
MAB_4723c|M.abscessus_ATCC_199 MLN----------STADAPRGQRFAESTVARALLDLFNPAQLDVLIR---
MAV_1646|M.avium_104 VVTVLRGVQPLLGQDPVIVQGFRAWAGSISPAAARFIEEIARQTARRGLA
Mflv_0932|M.gilvum_PYR-GCK ------IFGATP--PPQ---
MSMEG_3274|M.smegmatis_MC2_155 ------ILATSPSQPPVP--
Mvan_3507|M.vanbaalenii_PYR-1 ------VFRLAAGEPSAQK-
Mb3367|M.bovis_AF2122/97 --------------------
Rv3334|M.tuberculosis_H37Rv --------------------
MAB_4723c|M.abscessus_ATCC_199 ------VGAAVRSEPGEG--
MAV_1646|M.avium_104 AVSVIVLPAADRAQTADATP