For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SQTSRPPLIRATTHWLRTTVLAVAVLLIAAACTSSPAAAPTPDVITDKGTPFADLLVPKLQASVTDGAIG VAVDSPVTVSAGDGVLGQVSLTNDAGKIVDGQLSPDGVSWASAEPLGYNKQYTLQAQAHGLGGTTSTSAT FETHSPENLTMPYVLPNDGETVGVGQPIAIRFDENITNRVAAQQAITVTTKPPVEGAFYWLSNREVRWRP AEYWEPGTSVEVSVKAYGVDFGDGLFGQDDVTTRFTIGDQIIATADDATKTMTVRRNGDVVKTMPISMGK AKTPTDNGAYIIGDRYSFLVMDSSTYGVPVNSPDGYRTEVEWATQMSYSGIYVHSAPWSVGSQGIANVSH GCLNVNPANARWFYDNTRRGDIVEVINTTGATLSGTDGLGDWNIPWEQWKAGNANL*
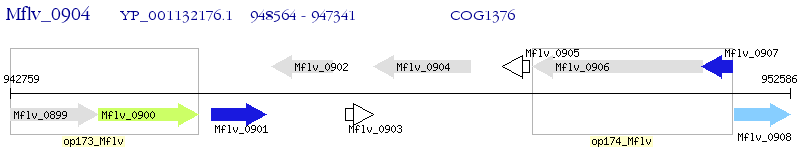
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0904 | - | - | 100% (407) | ErfK/YbiS/YcfS/YnhG family protein |
| M. gilvum PYR-GCK | Mflv_2542 | - | e-175 | 73.66% (391) | ErfK/YbiS/YcfS/YnhG family protein |
| M. gilvum PYR-GCK | Mflv_2844 | - | e-150 | 97.66% (256) | ErfK/YbiS/YcfS/YnhG family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2547c | lppS | 1e-162 | 66.33% (395) | lipoprotein LppS |
| M. tuberculosis H37Rv | Rv2518c | lppS | 1e-162 | 66.33% (395) | lipoprotein LppS |
| M. leprae Br4923 | MLBr_00426 | lppS | 1e-160 | 65.21% (388) | putative secreted protein |
| M. abscessus ATCC 19977 | MAB_1530 | - | 1e-158 | 64.21% (394) | lipoprotein LppS |
| M. marinum M | MMAR_3872 | lppS | 1e-163 | 66.42% (399) | lipoprotein LppS |
| M. avium 104 | MAV_1661 | - | 1e-163 | 69.50% (377) | ErfK/YbiS/YcfS/YnhG family protein |
| M. smegmatis MC2 155 | MSMEG_4745 | - | 1e-171 | 71.07% (401) | ErfK/YbiS/YcfS/YnhG family protein |
| M. thermoresistible (build 8) | TH_1098 | lppS | 1e-172 | 70.15% (392) | PROBABLE CONSERVED LIPOPROTEIN LPPS |
| M. ulcerans Agy99 | MUL_3804 | lppS | 1e-162 | 66.17% (399) | lipoprotein LppS |
| M. vanbaalenii PYR-1 | Mvan_3651 | - | 0.0 | 93.86% (407) | ErfK/YbiS/YcfS/YnhG family protein |
CLUSTAL 2.0.9 multiple sequence alignment
MLBr_00426|M.leprae_Br4923 ---MGTATRRVQPKAWRALLTLLVISVVMP--GVACN-RGGGNVPVNVIG
MAV_1661|M.avium_104 ----------------------------MG--AVACG-GGHTAAPPKVIF
Mb2547c|M.bovis_AF2122/97 MPKVGIAAQAGRTRVRRAWLTALMMTAVMIG-AVACG-SGRGPAPIKVIA
Rv2518c|M.tuberculosis_H37Rv MPKVGIAAQAGRTRVRRAWLTALMMTAVMIG-AVACG-SGRGPAPIKVIA
MMAR_3872|M.marinum_M ---MAPLSRR------RSWLAAILIPVAVL--AVACGGSSATPTATKVIE
MUL_3804|M.ulcerans_Agy99 --MMAPLSRR------RSWLAAILIPVAVL--AVACGGSSATPTATKVIE
MAB_1530|M.abscessus_ATCC_1997 ---MTQGRPRLHAGARRRWVATLALPVVAM--AVLAGCAGATTQEPPKVI
Mflv_0904|M.gilvum_PYR-GCK -MSQTSRPPLIRATTHWLRTTVLAVAVLLIA--AACTSSPAAAPTPDVIT
Mvan_3651|M.vanbaalenii_PYR-1 -MPQIGCPLPFRATTHWLRTTVLAVAVLLIA--TACTSSPAAAPTPDVIT
MSMEG_4745|M.smegmatis_MC2_155 ---------MIRPRRSLFRSAVVAMVPALLFGLMACGKDEQDGPAP-VIA
TH_1098|M.thermoresistible__bu ------MTRPRWRSRWWLITGLLSVVVLALS---ACSGG-GASDQPKVIT
. :
MLBr_00426|M.leprae_Br4923 DKGTPFADLLVPKLTASVTDGAVGVNVDMPVTVTVADGVLAAVTMINDNG
MAV_1661|M.avium_104 DKGTPFADLLVPKLTASVSDGAVGVTVDAPVTVSVADGVLASVTMVNENG
Mb2547c|M.bovis_AF2122/97 DKGTPFADLLVPKLTASVTDGAVGVTVDAPVSVTAADGVLAAVTMVNDNG
Rv2518c|M.tuberculosis_H37Rv DKGTPFADLLVPKLTASVTDGAVGVTVDAPVSVTAADGVLAAVTMVNDNG
MMAR_3872|M.marinum_M NKGTPFADLLVPKLTASVTDGAVGVTVDAPVTVSVSDGVLAAVTMVNESG
MUL_3804|M.ulcerans_Agy99 NKGTPFADLLVPKLTASVTDGAVGVTVDAPVTVSVSDGVLAAVTMVNESG
MAB_1530|M.abscessus_ATCC_1997 DKATPYADLLVPKLAMSVKDGAVGVAVDAPVTVTAGEGVLGSVTMVNSDG
Mflv_0904|M.gilvum_PYR-GCK DKGTPFADLLVPKLQASVTDGAIGVAVDSPVTVSAGDGVLGQVSLTNDAG
Mvan_3651|M.vanbaalenii_PYR-1 DKGTPYADLLVPKLEASVTDGAIGVAVDAPVTVTAGDGVLGQVSLTNEAG
MSMEG_4745|M.smegmatis_MC2_155 DKGTPFGDLLVPRVNASVTDGAVGVTVESPVTVTAENGVLGGVSMTDESG
TH_1098|M.thermoresistible__bu DKGTPYGDLLVPRVTASVTDKAVGVPVDEPVTVSAENGVLGAVTMVNEQG
:*.**:.*****:: **.* *:** *: **:*:. :***. *:: :. *
MLBr_00426|M.leprae_Br4923 RMINGQFSPDGLRWSTTEPLGFNRCYTLSAKALGLGGVVNRQMTFQTSSP
MAV_1661|M.avium_104 RSISGQLSPDGLRWSTTEQLGYNRRYTLTAKATGLGGAASKQMTFETSSP
Mb2547c|M.bovis_AF2122/97 RPVAGRLSPDGLRWSTTEQLGYNRRYTLNATALGLGGAATRQLTFQTSSP
Rv2518c|M.tuberculosis_H37Rv RPVAGRLSPDGLRWSTTEQLGYNRRYTLNATALGLGGAATRQLTFQTSSP
MMAR_3872|M.marinum_M KSISGKLSSDGLHWSTDEPLGYNRRYTLNAKALGLGGAATRQMTFQTSSP
MUL_3804|M.ulcerans_Agy99 KSISGKLSFDGLHWSTDEPLGYNRRYTLNAKALGLGGAATRQMTFQTSSP
MAB_1530|M.abscessus_ATCC_1997 KEIAGEIGPDGVTWTTTEPLGYDKQYTINADARGLGGVARANATFRTQSP
Mflv_0904|M.gilvum_PYR-GCK KIVDGQLSPDGVSWASAEPLGYNKQYTLQAQAHGLGGTTSTSATFETHSP
Mvan_3651|M.vanbaalenii_PYR-1 ELVDGQLSPDGVTWSSAEPLGYNKQYTLQAHAQGLGGTTSTSATFETHSP
MSMEG_4745|M.smegmatis_MC2_155 SPVAGTLSPDGLTWSTTDPLDYNEQYTLVAQSLGLGGVTNRRIQFETHSP
TH_1098|M.thermoresistible__bu RTVEGEFSEDGVSWSTTEPLGYNRQYTITAQSLGLGGVTTREMTFETHSP
: * :. **: *:: : *.::. **: * : ****.. *.* **
MLBr_00426|M.leprae_Br4923 AHLTMPYVNPGNGEIVGIGEPVAIRFDENIANRLAAQKAITITANPPVEG
MAV_1661|M.avium_104 AHLTMPYVSPADGEVVGVGEPVAIRFDENIANRAAAQKAITITTNPPVEG
Mb2547c|M.bovis_AF2122/97 AHLTMPYVMPGDGEVVGVGEPVAIRFDENIADRGAAEKAIKITTNPPVEG
Rv2518c|M.tuberculosis_H37Rv AHLTMPYVMPGDGEVVGVGEPVAIRFDENIADRGAAEKAIKITTNPPVEG
MMAR_3872|M.marinum_M AHLTMPYVSPADGEVVGVGEPVAIRFDENIANRLAAEKAIKITTKPAVEG
MUL_3804|M.ulcerans_Agy99 AHLTMPYVSPADGEVVGVGEPVAIRFDENIANRLAAEKAIKITTKPAVEG
MAB_1530|M.abscessus_ATCC_1997 DNMTMPYVMPGDGEVVGVGQTVAIRFDENIPNRAAAEKAIKITTNPPVEG
Mflv_0904|M.gilvum_PYR-GCK ENLTMPYVLPNDGETVGVGQPIAIRFDENITNRVAAQQAITVTTKPPVEG
Mvan_3651|M.vanbaalenii_PYR-1 ENLTMPYVLPNDGETVGVGQPIAIRFDENITNRVAAQQAITVTTNPPVEG
MSMEG_4745|M.smegmatis_MC2_155 QNLTMPYVLPNNGEVVGVGQPIAIRFDENISDRVAAQRAITVKTTPPVEG
TH_1098|M.thermoresistible__bu ANLTMPYVVPSPGEVVGVGQPVAVRFDENIPNRAAAERAIKVTTNPPVEG
::***** * ** **:*:.:*:******.:* **::**.:.:.*.***
MLBr_00426|M.leprae_Br4923 AFYWLNNREVRWRPEHFWKSGTAVDVAVNTYGVDLGEGMFGEDNVKTHFT
MAV_1661|M.avium_104 AFYWLNNREVRWRPEHFWKSGTVIDVAVNTYGVDLGDGMFGEDNVKTHFT
Mb2547c|M.bovis_AF2122/97 AFYWLNNREVRWRPEHFWKPGTAVDVAVNTYGVDLGEGMFGEDNVQTHFT
Rv2518c|M.tuberculosis_H37Rv AFYWLNNREVRWRPEHFWKPGTAVDVAVNTYGVDLGEGMFGEDNVQTHFT
MMAR_3872|M.marinum_M AFYWLNNREVRWRPEHFWKSGTSVDVAVNTYGVDLGEGMFGEDNVKTHFT
MUL_3804|M.ulcerans_Agy99 AFYWLNNREVRWRPEHFWKSGTSVDVAVNTYGVDLGEGMFGEDNVKTHFT
MAB_1530|M.abscessus_ATCC_1997 AFYWLNNREVRWRPESFWDSGTSVDVKVNTYGVNLGDGVFGQDNVASHFT
Mflv_0904|M.gilvum_PYR-GCK AFYWLSNREVRWRPAEYWEPGTSVEVSVKAYGVDFGDGLFGQDDVTTRFT
Mvan_3651|M.vanbaalenii_PYR-1 AFYWLSNREVRWRPAEYWEPGTHVEVSVEAYGVDFGDGLFGQEDVTTRFT
MSMEG_4745|M.smegmatis_MC2_155 AFYWLNNREVRWRPAEYWKPGTTVDVQVNTYGVELGDGLYGQDNVETHFK
TH_1098|M.thermoresistible__bu AFYWLNNREVRWRPASYWKPGTTVEVEVNTYGVDLGDGLFGQRNTKTHFT
*****.******** :*..** ::* *::***::*:*::*: :. ::*.
MLBr_00426|M.leprae_Br4923 IGDEVFATADDATKMLTVRVNGEVVKIMPTSMGKDSTPTANGIYIVGARF
MAV_1661|M.avium_104 IGDEVISTADDTTKTVTVRVNGEVVKTMPTSMGKDSTPTANGIYIVGARF
Mb2547c|M.bovis_AF2122/97 IGDEVIATADDNTKILTVRVNGEVVKSMPTSMGKDSTPTANGIYIVGSRY
Rv2518c|M.tuberculosis_H37Rv IGDEVIATADDNTKILTVRVNGEVVKSMPTSMGKDSTPTANGIYIVGSRY
MMAR_3872|M.marinum_M IGDEVIATADDNTKMLTIRVNGEVVKTMPTSMGKDSTPTANGIYIVGARF
MUL_3804|M.ulcerans_Agy99 IGDEVIATADDNTKMLTILVNGEVVKTMPTSMGKDSTPTANGIYIVGARF
MAB_1530|M.abscessus_ATCC_1997 IGDAVISRVDDTNKILNIERNGEIIKTMPTSMGKDKAPTNNGTYIIGERF
Mflv_0904|M.gilvum_PYR-GCK IGDQIIATADDATKTMTVRRNGDVVKTMPISMGKAKTPTDNGAYIIGDRY
Mvan_3651|M.vanbaalenii_PYR-1 IGDHIIATADDATKTMTVRRNGEVVKTMPISMGKAKTPTDNGAYIIGDRY
MSMEG_4745|M.smegmatis_MC2_155 IGDEVIAVADDNTKTLTVRRNGEVVKTMPISMGKNSTPTNNGVYIIGDRY
TH_1098|M.thermoresistible__bu IGDEVISTVDDNTKTLTVRRNGEVVKTMPVSMGKDSTPTNNGVYIVGDKR
*** ::: .** .* :.: **:::* ** **** .:** ** **:* :
MLBr_00426|M.leprae_Br4923 KHIIMDSSTYGVPVNSPNGYRADVDWATQLSYSGVFLHSAPWSVGAQGHT
MAV_1661|M.avium_104 KHIIMDSSTYGVPVNSPNGYRTEVDWATQMSYSGVFVHSAPWSVGAQGHT
Mb2547c|M.bovis_AF2122/97 KHIIMDSSTYGVPVNSPNGYRTDVDWATQISYSGVFVHSAPWSVGAQGHT
Rv2518c|M.tuberculosis_H37Rv KHIIMDSSTYGVPVNSPNGYRTDVDWATQISYSGVFVHSAPWSVGAQGHT
MMAR_3872|M.marinum_M KHIIMDSSTYGVPVNSPNGYRTDVNWATQLSYSGVYVHSAPWSVGAQGHT
MUL_3804|M.ulcerans_Agy99 KHIIMDSSTYGVPVNSPNGYRTDVNWATQLSYSGVYVHSAPWSVGAQGHT
MAB_1530|M.abscessus_ATCC_1997 KDLIMDSSTYGVAVNSPDGYRTKVQYATQMSYSGIYVHAAPWSVGAQGRT
Mflv_0904|M.gilvum_PYR-GCK SFLVMDSSTYGVPVNSPDGYRTEVEWATQMSYSGIYVHSAPWSVGSQGIA
Mvan_3651|M.vanbaalenii_PYR-1 SFLVMDSSTYGVPVNSPDGYRTEVEWATQMSYSGIYVHSAPWSVGSQGIA
MSMEG_4745|M.smegmatis_MC2_155 EHLVMDSSTYGVPVNSPNGYRTEVDYATQMSYSGIYVHGAPWSVGSQGYS
TH_1098|M.thermoresistible__bu AHMVMDSSTYGVPVNSPNGYRTEVDWAVQMSYSGIYVHSAPWSVGSQGYS
::********.****:***:.*::*.*:****:::*.******:** :
MLBr_00426|M.leprae_Br4923 NTSHGCLNVSPSNAQWFYDHVKRGDIVEVVNTVGDTLPGAEGLGDWNIPW
MAV_1661|M.avium_104 NTSHGCLNVSPSNAEWFYDHSKSGDIVEVVHTVGPTLPGTEGLGDWNIPW
Mb2547c|M.bovis_AF2122/97 NTSHGCLNVSPSNAQWFYDHVKRGDIVEVVNTVGGTLPGIDGLGDWNIPW
Rv2518c|M.tuberculosis_H37Rv NTSHGCLNVSPSNAQWFYDHVKRGDIVEVVNTVGGTLPGIDGLGDWNIPW
MMAR_3872|M.marinum_M NTSHGCLNVSPSNAEWFYDHIKTGDIVEVVNTVGGTLPGTEGLGDWNIPW
MUL_3804|M.ulcerans_Agy99 NTSHGCLNVSPSNAEWFYDHIKTGDIVEVVNTVGGTLPGTEGLGDWNIPW
MAB_1530|M.abscessus_ATCC_1997 NTSHGCLNVSTANAKWFYENTKRGDVVIVSNTVGPVLPGTEGLGDWNIPW
Mflv_0904|M.gilvum_PYR-GCK NVSHGCLNVNPANARWFYDNTRRGDIVEVINTTGATLSGTDGLGDWNIPW
Mvan_3651|M.vanbaalenii_PYR-1 NVSHGCLNVNPANARWFYDNTRRGDIVEVINTTGATLSGTDGLGDWNIPW
MSMEG_4745|M.smegmatis_MC2_155 NVSHGCLNVSTANARWFYENTKRGDIVEVRNTVGSLLPGTDGLGDWNIPW
TH_1098|M.thermoresistible__bu NVSHGCINVSPSNAKWFYENSKRGDIIEVVNTVGTVLPGTDGLGDWNIPW
*.****:**..:**.***:: : **:: * :*.* *.* :*********
MLBr_00426|M.leprae_Br4923 EQWKAGNANI-
MAV_1661|M.avium_104 SQWKAGNANT-
Mb2547c|M.bovis_AF2122/97 DQWRAGNAKA-
Rv2518c|M.tuberculosis_H37Rv DQWRAGNAKA-
MMAR_3872|M.marinum_M EQWRAGNANA-
MUL_3804|M.ulcerans_Agy99 EQWRAGNANA-
MAB_1530|M.abscessus_ATCC_1997 AQWKAGNARQQ
Mflv_0904|M.gilvum_PYR-GCK EQWKAGNANL-
Mvan_3651|M.vanbaalenii_PYR-1 EQWKAGNANL-
MSMEG_4745|M.smegmatis_MC2_155 EQWRAGNASI-
TH_1098|M.thermoresistible__bu EQWKAGNANV-
**:****