For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTQGRPRLHAGARRRWVATLALPVVAMAVLAGCAGATTQEPPKVIDKATPYADLLVPKLAMSVKDGAVGV AVDAPVTVTAGEGVLGSVTMVNSDGKEIAGEIGPDGVTWTTTEPLGYDKQYTINADARGLGGVARANATF RTQSPDNMTMPYVMPGDGEVVGVGQTVAIRFDENIPNRAAAEKAIKITTNPPVEGAFYWLNNREVRWRPE SFWDSGTSVDVKVNTYGVNLGDGVFGQDNVASHFTIGDAVISRVDDTNKILNIERNGEIIKTMPTSMGKD KAPTNNGTYIIGERFKDLIMDSSTYGVAVNSPDGYRTKVQYATQMSYSGIYVHAAPWSVGAQGRTNTSHG CLNVSTANAKWFYENTKRGDVVIVSNTVGPVLPGTEGLGDWNIPWAQWKAGNARQQ
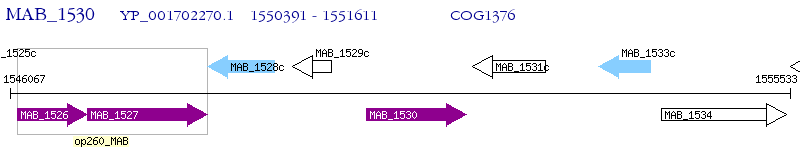
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_1530 | - | - | 100% (406) | lipoprotein LppS |
| M. abscessus ATCC 19977 | MAB_4537c | - | 1e-61 | 47.28% (239) | hypothetical protein MAB_4537c |
| M. abscessus ATCC 19977 | MAB_4061c | - | 7e-55 | 31.54% (409) | hypothetical protein MAB_4061c |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2547c | lppS | 1e-163 | 67.16% (402) | lipoprotein LppS |
| M. gilvum PYR-GCK | Mflv_2542 | - | 1e-158 | 63.84% (401) | ErfK/YbiS/YcfS/YnhG family protein |
| M. tuberculosis H37Rv | Rv2518c | lppS | 1e-163 | 67.16% (402) | lipoprotein LppS |
| M. leprae Br4923 | MLBr_00426 | lppS | 1e-156 | 65.31% (392) | putative secreted protein |
| M. marinum M | MMAR_3872 | lppS | 1e-163 | 68.54% (391) | lipoprotein LppS |
| M. avium 104 | MAV_1661 | - | 1e-164 | 71.77% (372) | ErfK/YbiS/YcfS/YnhG family protein |
| M. smegmatis MC2 155 | MSMEG_4745 | - | 1e-155 | 64.86% (387) | ErfK/YbiS/YcfS/YnhG family protein |
| M. thermoresistible (build 8) | TH_1098 | lppS | 1e-164 | 67.75% (400) | PROBABLE CONSERVED LIPOPROTEIN LPPS |
| M. ulcerans Agy99 | MUL_3804 | lppS | 1e-163 | 68.54% (391) | lipoprotein LppS |
| M. vanbaalenii PYR-1 | Mvan_3651 | - | 1e-159 | 64.72% (394) | ErfK/YbiS/YcfS/YnhG family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2547c|M.bovis_AF2122/97 MPKVGIAAQAGRTRVRRAWLTALMMTAVMIGAVACGSG-RGPAPIKVIAD
Rv2518c|M.tuberculosis_H37Rv MPKVGIAAQAGRTRVRRAWLTALMMTAVMIGAVACGSG-RGPAPIKVIAD
MLBr_00426|M.leprae_Br4923 ---MGTATRRVQPKAWRALLTLLVISVVMPG-VACNRG-GGNVPVNVIGD
MAV_1661|M.avium_104 -----------------------------MGAVACGGG-HTAAPPKVIFD
MMAR_3872|M.marinum_M ---MAPLSRR------RSWLAAILIPVAVLAVACGGSS-ATPTATKVIEN
MUL_3804|M.ulcerans_Agy99 --MMAPLSRR------RSWLAAILIPVAVLAVACGGSS-ATPTATKVIEN
MAB_1530|M.abscessus_ATCC_1997 ---MTQGRPRLHAGARRRWVATLALPVVAMAVLAGCAG-ATTQEPPKVID
Mflv_2542|M.gilvum_PYR-GCK --MGWVESAGRARWRTSGAVAAALVGAMVFGLTGCGGS-TEAPTPQVISD
MSMEG_4745|M.smegmatis_MC2_155 -----MIRPRRSLFRS---AVVAMVPALLFGLMACGKD-EQDGPAPVIAD
TH_1098|M.thermoresistible__bu --------MTRPRWRSRWWLITGLLSVVVLALSACSGG-GASDQPKVITD
Mvan_3651|M.vanbaalenii_PYR-1 --MPQIGCPLPFRATTHWLRTTVLAVAVLLIATACTSSPAAAPTPDVITD
. : :
Mb2547c|M.bovis_AF2122/97 KGTPFADLLVPKLTASVTDGAVGVTVDAPVSVTAADGVLAAVTMVNDNGR
Rv2518c|M.tuberculosis_H37Rv KGTPFADLLVPKLTASVTDGAVGVTVDAPVSVTAADGVLAAVTMVNDNGR
MLBr_00426|M.leprae_Br4923 KGTPFADLLVPKLTASVTDGAVGVNVDMPVTVTVADGVLAAVTMINDNGR
MAV_1661|M.avium_104 KGTPFADLLVPKLTASVSDGAVGVTVDAPVTVSVADGVLASVTMVNENGR
MMAR_3872|M.marinum_M KGTPFADLLVPKLTASVTDGAVGVTVDAPVTVSVSDGVLAAVTMVNESGK
MUL_3804|M.ulcerans_Agy99 KGTPFADLLVPKLTASVTDGAVGVTVDAPVTVSVSDGVLAAVTMVNESGK
MAB_1530|M.abscessus_ATCC_1997 KATPYADLLVPKLAMSVKDGAVGVAVDAPVTVTAGEGVLGSVTMVNSDGK
Mflv_2542|M.gilvum_PYR-GCK KGTPFGDLLVPKLNASVQDGAVGVSAGSPVTVSAEDGVLGAVSMVNENGK
MSMEG_4745|M.smegmatis_MC2_155 KGTPFGDLLVPRVNASVTDGAVGVTVESPVTVTAENGVLGGVSMTDESGS
TH_1098|M.thermoresistible__bu KGTPYGDLLVPRVTASVTDKAVGVPVDEPVTVSAENGVLGAVTMVNEQGR
Mvan_3651|M.vanbaalenii_PYR-1 KGTPYADLLVPKLEASVTDGAIGVAVDAPVTVTAGDGVLGQVSLTNEAGE
*.**:.*****:: ** * *:** . **:*:. :***. *:: :. *
Mb2547c|M.bovis_AF2122/97 PVAGRLSPDGLRWSTTEQLGYNRRYTLNATALGLGGAATRQLTFQTSSPA
Rv2518c|M.tuberculosis_H37Rv PVAGRLSPDGLRWSTTEQLGYNRRYTLNATALGLGGAATRQLTFQTSSPA
MLBr_00426|M.leprae_Br4923 MINGQFSPDGLRWSTTEPLGFNRCYTLSAKALGLGGVVNRQMTFQTSSPA
MAV_1661|M.avium_104 SISGQLSPDGLRWSTTEQLGYNRRYTLTAKATGLGGAASKQMTFETSSPA
MMAR_3872|M.marinum_M SISGKLSSDGLHWSTDEPLGYNRRYTLNAKALGLGGAATRQMTFQTSSPA
MUL_3804|M.ulcerans_Agy99 SISGKLSFDGLHWSTDEPLGYNRRYTLNAKALGLGGAATRQMTFQTSSPA
MAB_1530|M.abscessus_ATCC_1997 EIAGEIGPDGVTWTTTEPLGYDKQYTINADARGLGGVARANATFRTQSPD
Mflv_2542|M.gilvum_PYR-GCK AVEGELTEDGLTWETAEPLGYNKRYTLTAQALGLGGVTSRQMTFETHSPE
MSMEG_4745|M.smegmatis_MC2_155 PVAGTLSPDGLTWSTTDPLDYNEQYTLVAQSLGLGGVTNRRIQFETHSPQ
TH_1098|M.thermoresistible__bu TVEGEFSEDGVSWSTTEPLGYNRQYTITAQSLGLGGVTTREMTFETHSPA
Mvan_3651|M.vanbaalenii_PYR-1 LVDGQLSPDGVTWSSAEPLGYNKQYTLQAHAQGLGGTTSTSATFETHSPE
: * : **: * : : *.::. **: * : ****.. *.* **
Mb2547c|M.bovis_AF2122/97 HLTMPYVMPGDGEVVGVGEPVAIRFDENIADRGAAEKAIKITTNPPVEGA
Rv2518c|M.tuberculosis_H37Rv HLTMPYVMPGDGEVVGVGEPVAIRFDENIADRGAAEKAIKITTNPPVEGA
MLBr_00426|M.leprae_Br4923 HLTMPYVNPGNGEIVGIGEPVAIRFDENIANRLAAQKAITITANPPVEGA
MAV_1661|M.avium_104 HLTMPYVSPADGEVVGVGEPVAIRFDENIANRAAAQKAITITTNPPVEGA
MMAR_3872|M.marinum_M HLTMPYVSPADGEVVGVGEPVAIRFDENIANRLAAEKAIKITTKPAVEGA
MUL_3804|M.ulcerans_Agy99 HLTMPYVSPADGEVVGVGEPVAIRFDENIANRLAAEKAIKITTKPAVEGA
MAB_1530|M.abscessus_ATCC_1997 NMTMPYVMPGDGEVVGVGQTVAIRFDENIPNRAAAEKAIKITTNPPVEGA
Mflv_2542|M.gilvum_PYR-GCK NLTMPYVLPNDGEVVGVGQPVAIRFDENIPNRLAAQRAITVKTTPEVEGA
MSMEG_4745|M.smegmatis_MC2_155 NLTMPYVLPNNGEVVGVGQPIAIRFDENISDRVAAQRAITVKTTPPVEGA
TH_1098|M.thermoresistible__bu NLTMPYVVPSPGEVVGVGQPVAVRFDENIPNRAAAERAIKVTTNPPVEGA
Mvan_3651|M.vanbaalenii_PYR-1 NLTMPYVLPNDGETVGVGQPIAIRFDENITNRVAAQQAITVTTNPPVEGA
::***** * ** **:*:.:*:******.:* **::**.:.:.* ****
Mb2547c|M.bovis_AF2122/97 FYWLNNREVRWRPEHFWKPGTAVDVAVNTYGVDLGEGMFGEDNVQTHFTI
Rv2518c|M.tuberculosis_H37Rv FYWLNNREVRWRPEHFWKPGTAVDVAVNTYGVDLGEGMFGEDNVQTHFTI
MLBr_00426|M.leprae_Br4923 FYWLNNREVRWRPEHFWKSGTAVDVAVNTYGVDLGEGMFGEDNVKTHFTI
MAV_1661|M.avium_104 FYWLNNREVRWRPEHFWKSGTVIDVAVNTYGVDLGDGMFGEDNVKTHFTI
MMAR_3872|M.marinum_M FYWLNNREVRWRPEHFWKSGTSVDVAVNTYGVDLGEGMFGEDNVKTHFTI
MUL_3804|M.ulcerans_Agy99 FYWLNNREVRWRPEHFWKSGTSVDVAVNTYGVDLGEGMFGEDNVKTHFTI
MAB_1530|M.abscessus_ATCC_1997 FYWLNNREVRWRPESFWDSGTSVDVKVNTYGVNLGDGVFGQDNVASHFTI
Mflv_2542|M.gilvum_PYR-GCK FYWLNNREVRWRPAKYWKPGTTVEVEVNTYGVDLGDGLFGQDNVSTSFTI
MSMEG_4745|M.smegmatis_MC2_155 FYWLNNREVRWRPAEYWKPGTTVDVQVNTYGVELGDGLYGQDNVETHFKI
TH_1098|M.thermoresistible__bu FYWLNNREVRWRPASYWKPGTTVEVEVNTYGVDLGDGLFGQRNTKTHFTI
Mvan_3651|M.vanbaalenii_PYR-1 FYWLSNREVRWRPAEYWEPGTHVEVSVEAYGVDFGDGLFGQEDVTTRFTI
****.******** :*..** ::* *::***::*:*::*: :. : *.*
Mb2547c|M.bovis_AF2122/97 GDEVIATADDNTKILTVRVNGEVVKSMPTSMGKDSTPTANGIYIVGSRYK
Rv2518c|M.tuberculosis_H37Rv GDEVIATADDNTKILTVRVNGEVVKSMPTSMGKDSTPTANGIYIVGSRYK
MLBr_00426|M.leprae_Br4923 GDEVFATADDATKMLTVRVNGEVVKIMPTSMGKDSTPTANGIYIVGARFK
MAV_1661|M.avium_104 GDEVISTADDTTKTVTVRVNGEVVKTMPTSMGKDSTPTANGIYIVGARFK
MMAR_3872|M.marinum_M GDEVIATADDNTKMLTIRVNGEVVKTMPTSMGKDSTPTANGIYIVGARFK
MUL_3804|M.ulcerans_Agy99 GDEVIATADDNTKMLTILVNGEVVKTMPTSMGKDSTPTANGIYIVGARFK
MAB_1530|M.abscessus_ATCC_1997 GDAVISRVDDTNKILNIERNGEIIKTMPTSMGKDKAPTNNGTYIIGERFK
Mflv_2542|M.gilvum_PYR-GCK GDQMVTTVDDSTKTLTVRRNGEVVKTMPVSMGKNSTPTNNGVYIVGDRRS
MSMEG_4745|M.smegmatis_MC2_155 GDEVIAVADDNTKTLTVRRNGEVVKTMPISMGKNSTPTNNGVYIIGDRYE
TH_1098|M.thermoresistible__bu GDEVISTVDDNTKTLTVRRNGEVVKTMPVSMGKDSTPTNNGVYIVGDKRA
Mvan_3651|M.vanbaalenii_PYR-1 GDHIIATADDATKTMTVRRNGEVVKTMPISMGKAKTPTDNGAYIIGDRYS
** :.: .** .* :.: ***::* ** **** .:** ** **:* :
Mb2547c|M.bovis_AF2122/97 HIIMDSSTYGVPVNSPNGYRTDVDWATQISYSGVFVHSAPWSVGAQGHTN
Rv2518c|M.tuberculosis_H37Rv HIIMDSSTYGVPVNSPNGYRTDVDWATQISYSGVFVHSAPWSVGAQGHTN
MLBr_00426|M.leprae_Br4923 HIIMDSSTYGVPVNSPNGYRADVDWATQLSYSGVFLHSAPWSVGAQGHTN
MAV_1661|M.avium_104 HIIMDSSTYGVPVNSPNGYRTEVDWATQMSYSGVFVHSAPWSVGAQGHTN
MMAR_3872|M.marinum_M HIIMDSSTYGVPVNSPNGYRTDVNWATQLSYSGVYVHSAPWSVGAQGHTN
MUL_3804|M.ulcerans_Agy99 HIIMDSSTYGVPVNSPNGYRTDVNWATQLSYSGVYVHSAPWSVGAQGHTN
MAB_1530|M.abscessus_ATCC_1997 DLIMDSSTYGVAVNSPDGYRTKVQYATQMSYSGIYVHAAPWSVGAQGRTN
Mflv_2542|M.gilvum_PYR-GCK HMVMDSSTYGVPVNSANGYRTEVDWATQISYSGIYVHAAPWSVGSQGESN
MSMEG_4745|M.smegmatis_MC2_155 HLVMDSSTYGVPVNSPNGYRTEVDYATQMSYSGIYVHGAPWSVGSQGYSN
TH_1098|M.thermoresistible__bu HMVMDSSTYGVPVNSPNGYRTEVDWAVQMSYSGIYVHSAPWSVGSQGYSN
Mvan_3651|M.vanbaalenii_PYR-1 FLVMDSSTYGVPVNSPDGYRTEVEWATQMSYSGIYVHSAPWSVGSQGIAN
::********.***.:***:.*::*.*:****:::*.******:** :*
Mb2547c|M.bovis_AF2122/97 TSHGCLNVSPSNAQWFYDHVKRGDIVEVVNTVGGTLPGIDGLGDWNIPWD
Rv2518c|M.tuberculosis_H37Rv TSHGCLNVSPSNAQWFYDHVKRGDIVEVVNTVGGTLPGIDGLGDWNIPWD
MLBr_00426|M.leprae_Br4923 TSHGCLNVSPSNAQWFYDHVKRGDIVEVVNTVGDTLPGAEGLGDWNIPWE
MAV_1661|M.avium_104 TSHGCLNVSPSNAEWFYDHSKSGDIVEVVHTVGPTLPGTEGLGDWNIPWS
MMAR_3872|M.marinum_M TSHGCLNVSPSNAEWFYDHIKTGDIVEVVNTVGGTLPGTEGLGDWNIPWE
MUL_3804|M.ulcerans_Agy99 TSHGCLNVSPSNAEWFYDHIKTGDIVEVVNTVGGTLPGTEGLGDWNIPWE
MAB_1530|M.abscessus_ATCC_1997 TSHGCLNVSTANAKWFYENTKRGDVVIVSNTVGPVLPGTEGLGDWNIPWA
Mflv_2542|M.gilvum_PYR-GCK VSHGCINVSTSNGKWFYDNSKRGDIVEIVNTVGSPLPGTDGLGDWNIPWE
MSMEG_4745|M.smegmatis_MC2_155 VSHGCLNVSTANARWFYENTKRGDIVEVRNTVGSLLPGTDGLGDWNIPWE
TH_1098|M.thermoresistible__bu VSHGCINVSPSNAKWFYENSKRGDIIEVVNTVGTVLPGTDGLGDWNIPWE
Mvan_3651|M.vanbaalenii_PYR-1 VSHGCLNVNPANARWFYDNTRRGDIVEVINTTGATLSGTDGLGDWNIPWE
.****:**..:*..***:: : **:: : :*.* *.* :*********
Mb2547c|M.bovis_AF2122/97 QWRAGNAKA-
Rv2518c|M.tuberculosis_H37Rv QWRAGNAKA-
MLBr_00426|M.leprae_Br4923 QWKAGNANI-
MAV_1661|M.avium_104 QWKAGNANT-
MMAR_3872|M.marinum_M QWRAGNANA-
MUL_3804|M.ulcerans_Agy99 QWRAGNANA-
MAB_1530|M.abscessus_ATCC_1997 QWKAGNARQQ
Mflv_2542|M.gilvum_PYR-GCK QWKAGNANL-
MSMEG_4745|M.smegmatis_MC2_155 QWRAGNASI-
TH_1098|M.thermoresistible__bu QWKAGNANV-
Mvan_3651|M.vanbaalenii_PYR-1 QWKAGNANL-
**:****