For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MASTPNMSTPPVMIAIDPHKRSWTAAAVDVRLQPLATIRVPVSRDGYRTLLRFARRWPDTAWAIEGASGL GAPLTVRLRQDGITVLDVPAKLAARVRLLSTGHGRKSDEADAVSVGVAAHTSSSLNTAEIDSATMALRAL VEHRDDLVKTRTQTINRLHTVLTHLIPGGASRDLNAERAAELLRTVRPRDTAGKTLRALAADLISELRGL DRRIDKAVHDIEIAVKDCGTTLTELHGIGALTAAKIVSRVGSIQRFDSPAAFASYTGTAPIEVSSGDVVR HRLSRAGDRQLNYCLHVMAITQIRRDTPGRTYYLRKRANGKSHSEAMRCLKRRLSDTVYRQLLRDANTTT
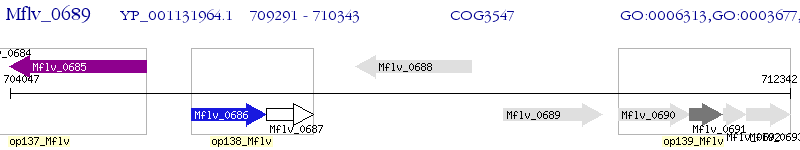
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0689 | - | - | 100% (350) | transposase IS116/IS110/IS902 family protein |
| M. gilvum PYR-GCK | Mflv_0450 | - | 4e-49 | 38.42% (341) | transposase IS116/IS110/IS902 family protein |
| M. gilvum PYR-GCK | Mflv_0001 | - | 8e-47 | 38.12% (341) | transposase IS116/IS110/IS902 family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3360 | - | 3e-43 | 37.09% (337) | transposase |
| M. tuberculosis H37Rv | Rv0797 | - | 3e-43 | 37.09% (337) | IS1547 transposase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_1466 | - | 1e-27 | 32.80% (311) | transposase |
| M. avium 104 | MAV_1059 | - | 1e-40 | 36.07% (341) | putative transposase |
| M. smegmatis MC2 155 | MSMEG_6023 | - | 3e-29 | 31.96% (341) | transposase |
| M. thermoresistible (build 8) | TH_3017 | - | 3e-19 | 28.11% (338) | PUTATIVE transposase |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0416 | - | 2e-17 | 26.15% (390) | transposase IS116/IS110/IS902 family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3360|M.bovis_AF2122/97 ----------MVVVGTDAHKYSHTFVATD-EVGRQLGEKTVKATTAGHAT
Rv0797|M.tuberculosis_H37Rv ----------MVVVGTDAHKYSHTFVATD-EVGRQLGEKTVKATTAGHAT
MAV_1059|M.avium_104 ----------MVVVGADVHKHTHTFVAVD-EVGRKLGEKTVKALTSGHAE
MMAR_1466|M.marinum_M --MTIVAHAHPFVIGVDTHARTHTLAVLVAATGEPVATEEFPTTAAGMNR
MSMEG_6023|M.smegmatis_MC2_155 -----MTPSQIVVIGADTHLDTIHLAALS-ETGKPLGDAEFPTRPSGYYV
Mflv_0689|M.gilvum_PYR-GCK MASTPNMSTPPVMIAIDPHKRSWTAAAVD-VRLQPLATIRVPVSRDGYRT
TH_3017|M.thermoresistible__bu -------------------------VVID-AQGKRLLSQRVANDEATLLK
Mvan_0416|M.vanbaalenii_PYR-1 ------MTTGQVWAGVDVGKEHHWVCAVD-DSGKVVLSRRLVNDEQPIRE
. . : .
Mb3360|M.bovis_AF2122/97 AIMWAREQFG--LELIWGIEDCRNMSARLERDLLAAGQQVVRVPTKLMAQ
Rv0797|M.tuberculosis_H37Rv AIMWAREQFG--LELIWGIEDCRNMSARLERDLLAAGQQVVRVPTKLMAQ
MAV_1059|M.avium_104 AVMWARERFG--AQVVWAIEDCRHLSARLERDLLTAGQQVVRVPPKLMAQ
MMAR_1466|M.marinum_M AVAWAARHTDGEMATLWVIEGIATYGAHLASAVKRAGFEVVEAA----AM
MSMEG_6023|M.smegmatis_MC2_155 AVKWAQSFGT---VTLAGVEGTNSYGAGLTRALQDGGIDVVEVN----RP
Mflv_0689|M.gilvum_PYR-GCK LLRFARRWPD----TAWAIEGASGLGAPLTVRLRQDGITVLDVPAKLAAR
TH_3017|M.thermoresistible__bu LISTVATMADGG-EVTWAIDLNAGGAALLITLLIAADQRLLYIPGRTVHH
Mvan_0416|M.vanbaalenii_PYR-1 LVAEIDELAER---VSWTVDLTTVYAALVLTVLADAGKTVRYLAGRAVWQ
: :: .* : : . :
Mb3360|M.bovis_AF2122/97 TRKSARSRG-KSDPIDALAVARAVLRE---TDLPLATHDETSRELKLLTD
Rv0797|M.tuberculosis_H37Rv TRKSARSRG-KSDPIDALAVARAVMRE---TDLPLATHDETSRELKLLTD
MAV_1059|M.avium_104 ARASARTRG-KSDPIDALAVARGFLRE---PDLPVASHDKVSRELKLLVD
MMAR_1466|M.marinum_M NARAHRGTG-KSDAFDARRIAASVLSLGPEQLRRPRSDDGIRAALRVLVT
MSMEG_6023|M.smegmatis_MC2_155 DRAARRRRG-KSDPLDAYAAARTALSG--HGLAAPK--DERTLALSALLT
Mflv_0689|M.gilvum_PYR-GCK VRLLSTGHGRKSDEADAVSVGVAAHTS---SSLNTAEIDSATMALRALVE
TH_3017|M.thermoresistible__bu ASGSYRG-EGKTDAKDAAVIADQARMR--RDLQPLRPGDDITVELRILTS
Mvan_0416|M.vanbaalenii_PYR-1 ASATYRGGEAKTDAKDARVIADQARMRG-QDLPVLHPDDDLISELRMLTG
*:* ** . * * *
Mb3360|M.bovis_AF2122/97 RRDVLVAQRTSAINRLRWLVHELDPERAPAARSLDAAKHQQALRTW----
Rv0797|M.tuberculosis_H37Rv RRDVLVAQRTSAINRLRWLVHELDPERAPAARSLDAAKHQQALRTW----
MAV_1059|M.avium_104 RREVLVAQRTAMINRLRWRVHELDPERAPGPASLDRGKHRHLLGAW----
MMAR_1466|M.marinum_M ARAHMTTERTATINALTALLRIA--ALGIDARKPLTASQIGEVARW----
MSMEG_6023|M.smegmatis_MC2_155 ARRGAVKAHTAATNQIQSLLVTAPSELRECYRRYTTRGLVKELARC----
Mflv_0689|M.gilvum_PYR-GCK HRDDLVKTRTQTINRLHTVLTHLIPGGASRDLNAERAAELLRTVRP----
TH_3017|M.thermoresistible__bu RRTDLVADRTRTINRLRAQLLEYFPALERAFDYSTSKAALLLLTGYQTPD
Mvan_0416|M.vanbaalenii_PYR-1 HRADLVADRTRTINRLRQQLVAVCPALERAAQPSQDRGWVILLARYQRPK
* . :* * : :
Mb3360|M.bovis_AF2122/97 -------------------LDTQPGLVAELARAELTDIIR-----LTGEI
Rv0797|M.tuberculosis_H37Rv -------------------LDTQPGLVAELARAELTDIIR-----LTGEI
MAV_1059|M.avium_104 -------------------LATVPGLVAELARDELADITR-----LTEAI
MMAR_1466|M.marinum_M -------------------RTRAEDIVTSTARAEAARMAKRVVDLNEQLA
MSMEG_6023|M.smegmatis_MC2_155 -------------------RPTAHTDPTAVAVLTALKALAYRAQFLHHQE
Mflv_0689|M.gilvum_PYR-GCK -------------------RDTAGKTLRALAADLISELRG-----LDRRI
TH_3017|M.thermoresistible__bu GLRRAGAARLAVWLGKRKARNADAVAAKALQAAHAQHTVIPGQQLAAAMV
Mvan_0416|M.vanbaalenii_PYR-1 AIRQSGVSRLTKVLTDAGVRNAASIAAAAVAAVKTQTMRLPGEEVAAGLI
Mb3360|M.bovis_AF2122/97 NTLAQRISARVHQVAP----------------ALLEIPGCAELTAAKIVG
Rv0797|M.tuberculosis_H37Rv NTLAQRISARVHQVAP----------------ALLEIPGCAELTAAKIVG
MAV_1059|M.avium_104 NALAKRIGSRVRTVAP----------------ALLAMPGCAELTAAKLVG
MMAR_1466|M.marinum_M ANQGQMIDLLHASKAA----------------ILLDKTGIGP-VTAAVVY
MSMEG_6023|M.smegmatis_MC2_155 HELTEQIHTLTQQMNP----------------GLRAAHGVGPDTAAALLI
Mflv_0689|M.gilvum_PYR-GCK DKAVHDIEIAVKDCGT----------------TLTELHGIGALTAAKIVS
TH_3017|M.thermoresistible__bu ARLAKEVMALDTEIGDTDAMIEERFHRHRHAEIIVSMSGFGVTLGAEFLA
Mvan_0416|M.vanbaalenii_PYR-1 ADLAREVIALDDRIKTTDADIEGRFRRHPLAEVITSMPGIGFRLGAEFLA
. : : * . * .:
Mb3360|M.bovis_AF2122/97 EAAG-VTRFKSEAAFACHAAVAPIPVWSGNTAGQMRLSRSGNRQLNAALH
Rv0797|M.tuberculosis_H37Rv EAAG-VTRFKSEAAFACHAAVAPIPVWSGNTAGQMRLSRSGNRQLNAALH
MAV_1059|M.avium_104 ETAG-VTRFKSEAAFARHAGVAPVPVWSGNTAGRVRMTRSGNRQLNAALH
MMAR_1466|M.marinum_M TAWSHAGRVRSEAAFAALAGVNPIPASSGNTI-RHRLNRGGDRRLNRALH
MSMEG_6023|M.smegmatis_MC2_155 TAGMNPHRLRSEAAFAALCGAAPVPASSGKTT-RHRLSRGGDRTANNALY
Mflv_0689|M.gilvum_PYR-GCK RVGS-IQRFDSPAAFASYTGTAPIEVSSGDVV-RHRLSRAGDRQLNYCLH
TH_3017|M.thermoresistible__bu ATGGDMSAFDSVDRLAGVAGLAPVPRDSGRISGNLQRPRRYNRRLLRVCY
Mvan_0416|M.vanbaalenii_PYR-1 AVG-DPALIGSADQLAAWAGLAPVSRDSGKRTGRLHTPKRYSRRLRRVMY
. . * :* . *: ** . : : .* :
Mb3360|M.bovis_AF2122/97 RIALTQIRMTDSRGQAYYQRLQDAGKTKRAALRCLKRRLARTVFQALRTV
Rv0797|M.tuberculosis_H37Rv RIALTQIRMTDSRGQAYYQRLQDAGKTKRAALRCLKRRLARTVFQALRTV
MAV_1059|M.avium_104 RIAITQIRLEG-LGQTYYRHRLAAGDSNPEALRCLKRRLARIIFGHLHTD
MMAR_1466|M.marinum_M MAVITRMTHDP-ETRA----------------------------------
MSMEG_6023|M.smegmatis_MC2_155 RIALVRMSNDP-RTPDYVARQTANGRSKIEIIRLLKRAVAREVFRLLSQP
Mflv_0689|M.gilvum_PYR-GCK VMAITQIRRDT-PGRTYYLRKRANGKSHSEAMRCLKRRLSDTVYRQLLRD
TH_3017|M.thermoresistible__bu LSALSSIRSDP-ASRTYYNRKRAEGKRHSQAVLALARRRLNVLWAMLRDH
Mvan_0416|M.vanbaalenii_PYR-1 MSALTAIRCDP-DSKAYYQRKRDEGKRPIPATICLARRRTNVLYALIRDN
.: :
Mb3360|M.bovis_AF2122/97 HQPSSEHTQPAAACHRSYCSSHLGEPPRLTDMTQKTRIQPLPPKRAGLLI
Rv0797|M.tuberculosis_H37Rv HQPSSEHTQPAAACHRSYCS-----RSCLSG-------------------
MAV_1059|M.avium_104 HQHRTQPCQTAAA-------------------------------------
MMAR_1466|M.marinum_M --------------------------------------------------
MSMEG_6023|M.smegmatis_MC2_155 CAIDDYSDLRPARQAKNITLTTVANHLDVWPNDISRLERGLKRDDTLAAN
Mflv_0689|M.gilvum_PYR-GCK ANTTT---------------------------------------------
TH_3017|M.thermoresistible__bu TTYQPTTPTAAAAA------------------------------------
Mvan_0416|M.vanbaalenii_PYR-1 RTWQPDSPPVTAAA------------------------------------
Mb3360|M.bovis_AF2122/97 RALYRIAKRRFGEVPEPFTVTAHHRRLLIANVVHEALLQRASRKLPPSVR
Rv0797|M.tuberculosis_H37Rv --------------------------------------------------
MAV_1059|M.avium_104 --------------------------------------------------
MMAR_1466|M.marinum_M --------------------------------------------------
MSMEG_6023|M.smegmatis_MC2_155 YREYLSAA------------------------------------------
Mflv_0689|M.gilvum_PYR-GCK --------------------------------------------------
TH_3017|M.thermoresistible__bu --------------------------------------------------
Mvan_0416|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb3360|M.bovis_AF2122/97 ELAVFWTARSIGCSWCVDFGAMLQRLDGLDVDRLTDIDNYATSSKFSDDE
Rv0797|M.tuberculosis_H37Rv --------------------------------------------------
MAV_1059|M.avium_104 --------------------------------------------------
MMAR_1466|M.marinum_M --------------------------------------------------
MSMEG_6023|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0689|M.gilvum_PYR-GCK --------------------------------------------------
TH_3017|M.thermoresistible__bu --------------------------------------------------
Mvan_0416|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb3360|M.bovis_AF2122/97 RAAIAYAEAMTADPHSVTDEQVADLRARFGEAGVIELTYQIGVENMRARM
Rv0797|M.tuberculosis_H37Rv --------------------------------------------------
MAV_1059|M.avium_104 --------------------------------------------------
MMAR_1466|M.marinum_M --------------------------------------------------
MSMEG_6023|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0689|M.gilvum_PYR-GCK --------------------------------------------------
TH_3017|M.thermoresistible__bu --------------------------------------------------
Mvan_0416|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb3360|M.bovis_AF2122/97 NSALGITEQGFNSGDACRVPWAAPDVPSAESR
Rv0797|M.tuberculosis_H37Rv --------------------------------
MAV_1059|M.avium_104 --------------------------------
MMAR_1466|M.marinum_M --------------------------------
MSMEG_6023|M.smegmatis_MC2_155 --------------------------------
Mflv_0689|M.gilvum_PYR-GCK --------------------------------
TH_3017|M.thermoresistible__bu --------------------------------
Mvan_0416|M.vanbaalenii_PYR-1 --------------------------------