For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TPSQIVVIGADTHLDTIHLAALSETGKPLGDAEFPTRPSGYYVAVKWAQSFGTVTLAGVEGTNSYGAGLT RALQDGGIDVVEVNRPDRAARRRRGKSDPLDAYAAARTALSGHGLAAPKDERTLALSALLTARRGAVKAH TAATNQIQSLLVTAPSELRECYRRYTTRGLVKELARCRPTAHTDPTAVAVLTALKALAYRAQFLHHQEHE LTEQIHTLTQQMNPGLRAAHGVGPDTAAALLITAGMNPHRLRSEAAFAALCGAAPVPASSGKTTRHRLSR GGDRTANNALYRIALVRMSNDPRTPDYVARQTANGRSKIEIIRLLKRAVAREVFRLLSQPCAIDDYSDLR PARQAKNITLTTVANHLDVWPNDISRLERGLKRDDTLAANYREYLSAA*
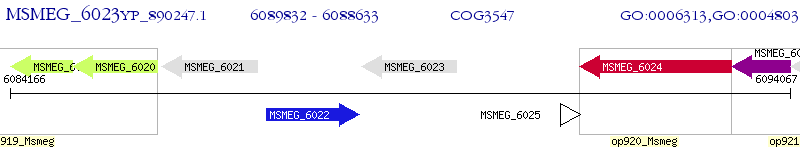
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. smegmatis MC2 155 | MSMEG_6023 | - | - | 100% (399) | transposase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3360 | - | 3e-31 | 33.43% (341) | transposase |
| M. gilvum PYR-GCK | Mflv_0450 | - | 1e-29 | 33.43% (359) | transposase IS116/IS110/IS902 family protein |
| M. tuberculosis H37Rv | Rv3327 | - | 3e-31 | 33.43% (341) | transposase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_3792 | - | 5e-50 | 42.77% (339) | transposase |
| M. avium 104 | MAV_1059 | - | 2e-31 | 34.43% (366) | putative transposase |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_4581 | - | 1e-05 | 27.50% (280) | transposase IS116/IS110/IS902 family protein |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_6023|M.smegmatis_MC2_155 ----MTPSQIV--VIGADTHLDTIHLAAL-SETGKPLGDAEFPTRPSGYY
MMAR_3792|M.marinum_M ----MIVAEAFGYVVGIDTHARTHTYCILNARTGAIVVGATFPNTSPAHR
Mb3360|M.bovis_AF2122/97 -----------MVVVGTDAHKYSHTFVAT-DEVGRQLGEKTVKATTAGHA
Rv3327|M.tuberculosis_H37Rv -----------MVVVGTDAHKYSHTFVAT-DEVGRQLGEKTVKATTAGHA
MAV_1059|M.avium_104 -----------MVVVGADVHKHTHTFVAV-DEVGRKLGEKTVKALTSGHA
Mflv_0450|M.gilvum_PYR-GCK -----------MVIVGTDVHKRTHTFVAV-DEVGRKLGEKVVPATTAGHR
Mvan_4581|M.vanbaalenii_PYR-1 MSVQCAPVTSSTIVVAVDVGKTAALFSVTDAARHLLVSPTEFTMNRSGLG
::. *. : : . ..
MSMEG_6023|M.smegmatis_MC2_155 VAVKWAQSFG---TVTLAGVEGTNSYGAGLTRALQDGGIDVVEVN----R
MMAR_3792|M.marinum_M RVIAWIQRHCPN-TKVLAAVEGTSSYGAGVTAALVDAGIDVTEV-----R
Mb3360|M.bovis_AF2122/97 TAIMWAREQFG--LELIWGIEDCRNMSARLERDLLAAGQQVVRVPTKLMA
Rv3327|M.tuberculosis_H37Rv TAIMWAREQFG--LELIWGIEDCRNMSARLERDLLAAGQQVVRVPTKLMA
MAV_1059|M.avium_104 EAVMWARERFG--AQVVWAIEDCRHLSARLERDLLTAGQQVVRVPPKLMA
Mflv_0450|M.gilvum_PYR-GCK TAVAWAREQFG--DELLWGIEDCRNLSARLEIDLLGMAQSAVRVAPKLMS
Mvan_4581|M.vanbaalenii_PYR-1 GAAASVMAAVPVSGQVKVAVEAAGHYHRPVLDHLWPDGWEVLELNPAHVA
. .:* : * . .. .:
MSMEG_6023|M.smegmatis_MC2_155 PDRAAR-RRRGKSDPLDAYAAARTALSG--HGLAAPKDE-RTLALSALLT
MMAR_3792|M.marinum_M PTGRHR-ARAGKSDVIDAEAAARSVLGCEQQRLPIPRESGEREDLRILLA
Mb3360|M.bovis_AF2122/97 QTRKSA-RSRGKSDPIDALAVARAVLRE--TDLPLATHDETSRELKLLTD
Rv3327|M.tuberculosis_H37Rv QTRKSA-RSRGKSDPIDALAVARAVLRE--TDLPLATHDETSRELKLLTD
MAV_1059|M.avium_104 QARASA-RTRGKSDPIDALAVARGFLRE--PDLPVASHDKVSRELKLLVD
Mflv_0450|M.gilvum_PYR-GCK QSRACA-RTRGKSDPIDALAVAHAVLRH--PDLPVAAHDEVSRELKLLTD
Mvan_4581|M.vanbaalenii_PYR-1 EQRRVQGRRRVKTDVIDLEAITELALSG--HGRAIADRDVVIGELSAWAG
*:* :* * :. * . . . *
MSMEG_6023|M.smegmatis_MC2_155 ARRGAVKAHTAATNQIQSLLVTAPSELRECYRRYTTRGLVKELARCRPTA
MMAR_3792|M.marinum_M ARRILDQQRTANRNALTALLRTI--DLGIDARKPLTNDQVRVIAAWRGNQ
Mb3360|M.bovis_AF2122/97 RRDVLVAQRTSAINRLRWLVHELDPERAPAARSLDAAKHQQALRTWLDTQ
Rv3327|M.tuberculosis_H37Rv RRDVLVAQRTSAINRLRWLVHELDPERAPAARSLDAAKHQQALRTWLDTQ
MAV_1059|M.avium_104 RREVLVAQRTAMINRLRWRVHELDPERAPGPASLDRGKHRHLLGAWLATV
Mflv_0450|M.gilvum_PYR-GCK RREDLVAQRTSTINRLRQRVHELDPAAEPNPGSLHRPTPCATLAAWLNTQ
Mvan_4581|M.vanbaalenii_PYR-1 HRSRRITTRTATKNQLLGQLDRAFPGLTLALPDVLGTKIGRLVAAEFSDP
* :*: * : : :
MSMEG_6023|M.smegmatis_MC2_155 HTDPTAVAVLTALKALAYRAQFLHH--QEHELTEQIHTLTQQMNPGLRAA
MMAR_3792|M.marinum_M KPSSTTAHAVARREARRLAKAVLEQTVQLDDNHRQLRGLAGQAAPGLQEL
Mb3360|M.bovis_AF2122/97 PG-------LVAELARAELTDIIRLTGEINTLAQRISARVHQVAPALLEI
Rv3327|M.tuberculosis_H37Rv PG-------LVAELARAELTDIIRLTGEINTLAQRISARVHQVAPALLEI
MAV_1059|M.avium_104 PG-------LVAELARDELADITRLTEAINALAKRIGSRVRTVAPALLAM
Mflv_0450|M.gilvum_PYR-GCK TG-------ILSELARDELADIVRLSEAINAVAARIGERVREVAPTLLAL
Mvan_4581|M.vanbaalenii_PYR-1 SR------LAALGVNRLIRFAAVRDLQLRRPVAERLVAAARDALP-TRDA
. :: . *
MSMEG_6023|M.smegmatis_MC2_155 HGVGPDTAAALLITAGMNPHRLRSEAAFAALCGAAPVPASSGKTT-RHRL
MMAR_3792|M.marinum_M PGIGP-VTAAIIICAYSHRGRIRSEAAFAALGGVAPLPASSGNTS-RHRL
Mb3360|M.bovis_AF2122/97 PGCAE-LTAAKIVGEAAGVTRFKSEAAFACHAAVAPIPVWSGNTAGQMRL
Rv3327|M.tuberculosis_H37Rv PGCAE-LTAAKIVGEAAGVTRFKSEAAFACHAAVAPIPVWSGNTAGQMRL
MAV_1059|M.avium_104 PGCAE-LTAAKLVGETAGVTRFKSEAAFARHAGVAPVPVWSGNTAGRVRM
Mflv_0450|M.gilvum_PYR-GCK PGCGE-LTAAKILGETATVSRFRSEAAFARHTGTAPIPVWSGNTAGRVRL
Mvan_4581|M.vanbaalenii_PYR-1 AIARQILAADLNLLAGLDAQIQVAEAALAVLLPRSPFSTLTTVPGWGVVR
:* : :***:* :*... : .
MSMEG_6023|M.smegmatis_MC2_155 SRGGDRTANNALYRIALVRMSNDP-RTPDYVA--RQTANGRSKIEIIRLL
MMAR_3792|M.marinum_M SRAGDRQLNRAFEVIVRTRMTCDP-ATRHYVA--RRTAEGMSKRETRRCL
Mb3360|M.bovis_AF2122/97 SRSGNRQLNAALHRIALTQIRMTDSRGQAYYQ--RLQDAGKTKRAALRCL
Rv3327|M.tuberculosis_H37Rv SRSGNRQLNAALHRIALTQIRMTDSRGQAYYQ--RLQDAGKTKRAALRCL
MAV_1059|M.avium_104 TRSGNRQLNAALHRIAITQIRLEG-LGQTYYR--HRLAAGDSNPEALRCL
Mflv_0450|M.gilvum_PYR-GCK SRSGNRQLNAALHRIAITQIRLTDSAGQRYYR--KRLAEGKATKEALRCL
Mvan_4581|M.vanbaalenii_PYR-1 AANYAAALGDPYRWPGPRQIYRASGLSPMQYESAHKRRDGGISREGSVAL
: . . :: : * . *
MSMEG_6023|M.smegmatis_MC2_155 KRAVAREVFRLLSQPCAIDDYSDLRPARQAKNITLTTVANHLDVWPNDIS
MMAR_3792|M.marinum_M KRYVCRSVFRHLQAATT---------------------------------
Mb3360|M.bovis_AF2122/97 KRRLARTVFQALRTVHQPSSEHTQPAAACHRSYCSSHLGEPPRLTDMTQK
Rv3327|M.tuberculosis_H37Rv KRRLARTVFQALRTVHQPSSEHTQPAAACHRSYCSSHLGEPPRLTDMTQK
MAV_1059|M.avium_104 KRRLARIIFGHLHTDHQHRTQPCQTAAA----------------------
Mflv_0450|M.gilvum_PYR-GCK KRQLARIVYRHLHTDENTRKTKPHNNAQTAA-------------------
Mvan_4581|M.vanbaalenii_PYR-1 RRALIDLGLGLWLTEPAAKAYAGGLKARGKRGGVIACALAHRATRIAHAL
:* :
MSMEG_6023|M.smegmatis_MC2_155 RLERGLKRDDTLAANYREYLSAA---------------------------
MMAR_3792|M.marinum_M --------------------------------------------------
Mb3360|M.bovis_AF2122/97 TRIQPLPPKRAGLLIRALYRIAKRRFGEVPEPFTVTAHHRRLLIANVVHE
Rv3327|M.tuberculosis_H37Rv TRIQPLPPKRAGLLIRALYRIAKRRFGEVPEPFTVTAHHRRLLIANVVHE
MAV_1059|M.avium_104 --------------------------------------------------
Mflv_0450|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4581|M.vanbaalenii_PYR-1 VRDHAAYDPARWI-------------------------------------
MSMEG_6023|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_3792|M.marinum_M --------------------------------------------------
Mb3360|M.bovis_AF2122/97 ALLQRASRKLPPSVRELAVFWTARSIGCSWCVDFGAMLQRLDGLDVDRLT
Rv3327|M.tuberculosis_H37Rv ALLQRASRKLPPSVRELAVFWTARSIGCSWCVDFGAMLQRLDGLDVDRLT
MAV_1059|M.avium_104 --------------------------------------------------
Mflv_0450|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4581|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6023|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_3792|M.marinum_M --------------------------------------------------
Mb3360|M.bovis_AF2122/97 DIDNYATSSKFSDDERAAIAYAEAMTADPHSVTDEQVADLRARFGEAGVI
Rv3327|M.tuberculosis_H37Rv DIDNYATSSKFSDDERAAIAYAEAMTADPHSVTDEQVADLRARFGEAGVI
MAV_1059|M.avium_104 --------------------------------------------------
Mflv_0450|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_4581|M.vanbaalenii_PYR-1 --------------------------------------------------
MSMEG_6023|M.smegmatis_MC2_155 -----------------------------------------------
MMAR_3792|M.marinum_M -----------------------------------------------
Mb3360|M.bovis_AF2122/97 ELTYQIGVENMRARMNSALGITEQGFNSGDACRVPWAAPDVPSAESR
Rv3327|M.tuberculosis_H37Rv ELTYQIGVENMRARMNSALGITEQGFNSGDACRVPWAAPDVPSAESR
MAV_1059|M.avium_104 -----------------------------------------------
Mflv_0450|M.gilvum_PYR-GCK -----------------------------------------------
Mvan_4581|M.vanbaalenii_PYR-1 -----------------------------------------------