For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TTGQVWAGVDVGKEHHWVCAVDDSGKVVLSRRLVNDEQPIRELVAEIDELAERVSWTVDLTTVYAALVLT VLADAGKTVRYLAGRAVWQASATYRGGEAKTDAKDARVIADQARMRGQDLPVLHPDDDLISELRMLTGHR ADLVADRTRTINRLRQQLVAVCPALERAAQPSQDRGWVILLARYQRPKAIRQSGVSRLTKVLTDAGVRNA ASIAAAAVAAVKTQTMRLPGEEVAAGLIADLAREVIALDDRIKTTDADIEGRFRRHPLAEVITSMPGIGF RLGAEFLAAVGDPALIGSADQLAAWAGLAPVSRDSGKRTGRLHTPKRYSRRLRRVMYMSALTAIRCDPDS KAYYQRKRDEGKRPIPATICLARRRTNVLYALIRDNRTWQPDSPPVTAAA*
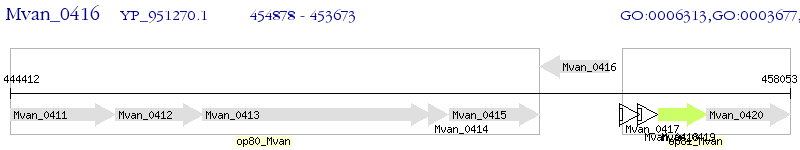
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. vanbaalenii PYR-1 | Mvan_0416 | - | - | 100% (401) | transposase IS116/IS110/IS902 family protein |
| M. vanbaalenii PYR-1 | Mvan_4491 | - | 1e-27 | 26.75% (400) | transposase, IS111A/IS1328/IS1533 |
| M. vanbaalenii PYR-1 | Mvan_5958 | - | 6e-27 | 26.50% (400) | transposase, IS111A/IS1328/IS1533 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3360 | - | 3e-18 | 25.87% (402) | transposase |
| M. gilvum PYR-GCK | Mflv_4200 | - | 0.0 | 100.00% (401) | transposase IS116/IS110/IS902 family protein |
| M. tuberculosis H37Rv | Rv0797 | - | 1e-18 | 26.12% (402) | IS1547 transposase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_3782 | - | 4e-85 | 45.41% (392) | transposase |
| M. avium 104 | MAV_0496 | - | 4e-22 | 28.41% (345) | transposase IS116/IS110/IS902 |
| M. smegmatis MC2 155 | MSMEG_4550 | - | 0.0 | 95.26% (401) | transposase |
| M. thermoresistible (build 8) | TH_3017 | - | 1e-99 | 52.22% (383) | PUTATIVE transposase |
| M. ulcerans Agy99 | - | - | - | - | - |
CLUSTAL 2.0.9 multiple sequence alignment
Mvan_0416|M.vanbaalenii_PYR-1 MTTGQVWAGVDVGKEHHWVCAVDDSGKVVLSRRLVNDEQPIRELVAEIDE
Mflv_4200|M.gilvum_PYR-GCK MTTGQVWAGVDVGKEHHWVCAVDDSGKVVLSRRLVNDEQPIRELVAEIDE
MSMEG_4550|M.smegmatis_MC2_155 MAAGQVWAGVDVGKEHHWVCVVDDSGKVVLSRRLVNDEQPIRELVAEIDE
MMAR_3782|M.marinum_M --MPTVWAGVDTGKRTHHCVVLDRNGGVLLSRPVDNDETALLELIEAVVG
TH_3017|M.thermoresistible__bu -------------------VVIDAQGKRLLSQRVANDEATLLKLISTVAT
Mb3360|M.bovis_AF2122/97 ----MVVVGTDAHKYSHTFVATDEVGR-QLGEKTVKATTAGHATAIMWAR
Rv0797|M.tuberculosis_H37Rv ----MVVVGTDAHKYSHTFVATDEVGR-QLGEKTVKATTAGHATAIMWAR
MAV_0496|M.avium_104 ---------------------------MVARGRVSNDAAGFAALLTLLAE
:
Mvan_0416|M.vanbaalenii_PYR-1 LAER--VSWTVDLTTVYAALVLTVLADAGKTVRYLAGRAVWQASATYRGG
Mflv_4200|M.gilvum_PYR-GCK LAER--VSWTVDLTTVYAALVLTVLADAGKTVRYLAGRAVWQASATYRGG
MSMEG_4550|M.smegmatis_MC2_155 LAER--VLWAVDLTTVYAALLLTVLADAGKTVRYLAGRAVWQASATYRGG
MMAR_3782|M.marinum_M VAAGREVCWATDLNAGGAAMLITLLAAHDQQLLYIPGRIVHHAAATYRG-
TH_3017|M.thermoresistible__bu MADGGEVTWAIDLNAGGAALLITLLIAADQRLLYIPGRTVHHASGSYRG-
Mb3360|M.bovis_AF2122/97 EQFGLELIWGIEDCRNMSARLERDLLAAGQQVVRVPTKLMAQTRKSARS-
Rv0797|M.tuberculosis_H37Rv EQFGLELIWGIEDCRNMSARLERDLLAAGQQVVRVPTKLMAQTRKSARS-
MAV_0496|M.avium_104 AGDSAAHPIPVGIETDR-GLWVAALRETGRVIYPINPLAASRYRARHAVS
. * .: : : :
Mvan_0416|M.vanbaalenii_PYR-1 EAKTDAKDARVIADQARMRGQDLPVLHPDDDLISELRMLTGHRADLVADR
Mflv_4200|M.gilvum_PYR-GCK EAKTDAKDARVIADQARMRGQDLPVLHPDDDLISELRMLTGHRADLVADR
MSMEG_4550|M.smegmatis_MC2_155 EAKTDAKDARVIADQSRMRGQDLPVLHPNDDLISELRMLTGHRADLVADR
MMAR_3782|M.marinum_M DGKTDAKNARIIADQARMR-TDLQPVRRADPIATDLRLLTSRRTDVVCDR
TH_3017|M.thermoresistible__bu EGKTDAKDAAVIADQARMR-RDLQPLRPGDDITVELRILTSRRTDLVADR
Mb3360|M.bovis_AF2122/97 RGKSDPIDALAVAR-AVLRETDLP-LATHDETSRELKLLTDRRDVLVAQR
Rv0797|M.tuberculosis_H37Rv RGKSDPIDALAVAR-AVMRETDLP-LATHDETSRELKLLTDRRDVLVAQR
MAV_0496|M.avium_104 GAKSDATDAVLLANIVRTDAAAHRPLPADTELAQAIRVLARAQQDAVWAR
.*:*. :* :* : :::*: : * *
Mvan_0416|M.vanbaalenii_PYR-1 TRTINRLRQQLVAVCPALERAAQPSQDRGWV-----ILLARYQRPKAIRQ
Mflv_4200|M.gilvum_PYR-GCK TRTINRLRQQLVAVCPALERAAQPSQDRGWV-----ILLARYQRPKAIRQ
MSMEG_4550|M.smegmatis_MC2_155 TRTINRLRQQLVAVCPALERAAQLSQDRGWV-----VLLARYQRPKAIRQ
MMAR_3782|M.marinum_M VRAINRLRQTLLEYFPPLERAFDYSKSKAAL-----TLLSRYQTPDSLRR
TH_3017|M.thermoresistible__bu TRTINRLRAQLLEYFPALERAFDYSTSKAAL-----LLLTGYQTPDGLRR
Mb3360|M.bovis_AF2122/97 TSAINRLRWLVHELDPERAPAAR---------------------------
Rv0797|M.tuberculosis_H37Rv TSAINRLRWLVHELDPERAPAAR---------------------------
MAV_0496|M.avium_104 QQIGNQIRDLLKDFYPAAIAAFAGLPEGGLARADARTILAAAPTPTQAAT
*::* : * *
Mvan_0416|M.vanbaalenii_PYR-1 SGVSRLTKVLTDAGVRNAASIAAAAVAAVKTQTMRLPGEEVAAGL---IA
Mflv_4200|M.gilvum_PYR-GCK SGVSRLTKVLTDAGVRNAASIAAAAVAAVKTQTMRLPGEEVAAGL---IA
MSMEG_4550|M.smegmatis_MC2_155 SGVSRLTKVLTDAGVRNAASIAGAAVAAVKTQTVRLPGEEVAAGL---VA
MMAR_3782|M.marinum_M IGPTRLATWLKARGCRNSATVAQTAVQAAQTQHTTLPTQTVGAQL---VP
TH_3017|M.thermoresistible__bu AGAARLAVWLGKRKARNADAVAAKALQAAHAQHTVIPGQQLAAAM---VA
Mb3360|M.bovis_AF2122/97 ------------------SLDAAKHQQALRTWLDTQPG--LVAEL---AR
Rv0797|M.tuberculosis_H37Rv ------------------SLDAAKHQQALRTWLDTQPG--LVAEL---AR
MAV_0496|M.avium_104 LTPARLRRLLTKAGRRRHLDRDIERLRAVFTDTYLHQPPAVENAMGIQLA
* : : :
Mvan_0416|M.vanbaalenii_PYR-1 DLAREVIALDDRIKTTDADIEGRFRRHPLAEVITSMPGIGFRLGAEFLAA
Mflv_4200|M.gilvum_PYR-GCK DLAREVIALDDRIKTTDADIEGRFRRHPLAEVITSMPGIGFRLGAEFLAA
MSMEG_4550|M.smegmatis_MC2_155 DLAREVIALDDRIKTTDADIEGRFRRHPLAEVITSMPGMGFRLGAELLAA
MMAR_3782|M.marinum_M RLAAEITTIDAELAHVDNEITARFADHDSAEILTSMPGFGPVLAATFLAQ
TH_3017|M.thermoresistible__bu RLAKEVMALDTEIGDTDAMIEERFHRHRHAEIIVSMSGFGVTLGAEFLAA
Mb3360|M.bovis_AF2122/97 AELTDIIRLTGEINTLAQRISARV--HQVAPALLEIPGCAELTAAKIVGE
Rv0797|M.tuberculosis_H37Rv AELTDIIRLTGEINTLAQRISARV--HQVAPALLEIPGCAELTAAKIVGE
MAV_0496|M.avium_104 ALLRQLEAACTAADELAEAAIAHFEQHPDAKIITSFPGLGMLAGARVLAE
:: :. * * : .:.* . .* .:.
Mvan_0416|M.vanbaalenii_PYR-1 VG-DPALIGSADQLAAWAGLAPVSRDSGKRTGRLHTPKRYSRRLRRVMYM
Mflv_4200|M.gilvum_PYR-GCK VG-DPALIGSADQLAAWAGLAPVSRDSGKRTGRLHTPKRYSRRLRRVMYM
MSMEG_4550|M.smegmatis_MC2_155 VG-DPALIGSADQLAAWAGLAPVSRDSRKRTGRLHTPKRYSRRLRRVMYM
MMAR_3782|M.marinum_M IGGNLDGFDTVDRLACVAGLAPVPRDSGRISGNLHRPRRFNRRLLRPCYL
TH_3017|M.thermoresistible__bu TGGDMSAFDSVDRLAGVAGLAPVPRDSGRISGNLQRPRRYNRRLLRVCYL
Mb3360|M.bovis_AF2122/97 AA-GVTRFKSEAAFACHAAVAPIPVWSGNTAGQMRLSRSGNRQLNAALHR
Rv0797|M.tuberculosis_H37Rv AA-GVTRFKSEAAFACHAAVAPIPVWSGNTAGQMRLSRSGNRQLNAALHR
MAV_0496|M.avium_104 IGDDRTRFADARGLKAFAGSAPITRASGKKTVVLHRHIK-NRRLAAVGPI
. . : : *. **:. * . : :: .*:*
Mvan_0416|M.vanbaalenii_PYR-1 SALTAIRC-DPDSKAYYQRKRDEGKRPIPATICLARRRTNVLYALIRDNR
Mflv_4200|M.gilvum_PYR-GCK SALTAIRC-DPDSKAYYQRKRDEGKRPIPATICLARRRTNVLYALIRDNR
MSMEG_4550|M.smegmatis_MC2_155 SALTAIRC-DPDSKAYYQRKRDEGKRPIPATICLARRRTNVLYALIRDNR
MMAR_3782|M.marinum_M AALSSLKN-SPASRTFYDRKRAEGKSHKQALIALARRRINVIWAMLRDHT
TH_3017|M.thermoresistible__bu SALSSIRS-DPASRTYYNRKRAEGKRHSQAVLALARRRLNVLWAMLRDHT
Mb3360|M.bovis_AF2122/97 IALTQIRMTDSRGQAYYQRLQDAGKTKRAALRCLKRRLARTVFQALRTVH
Rv0797|M.tuberculosis_H37Rv IALTQIRMTDSRGQAYYQRLQDAGKTKRAALRCLKRRLARTVFQALRTVH
MAV_0496|M.avium_104 WALGSLRA-SPGARRHYDARRAAGDWNHQAQRHLFNKFLGQLHHCLQTGR
** :: .. .: .*: : *. * * .: : ::
Mvan_0416|M.vanbaalenii_PYR-1 TWQPDSPPVTAAA-------------------------------------
Mflv_4200|M.gilvum_PYR-GCK TWQPDSPPVTAAA-------------------------------------
MSMEG_4550|M.smegmatis_MC2_155 TWQPESPPNIAAA-------------------------------------
MMAR_3782|M.marinum_M HYHEPDLVNTPLAA------------------------------------
TH_3017|M.thermoresistible__bu TYQPTTPTAAAAA-------------------------------------
Mb3360|M.bovis_AF2122/97 QPSSEHTQPAAACHRSYCSSHLGEPPRLTDMTQKTRIQPLPPKRAGLLIR
Rv0797|M.tuberculosis_H37Rv QPSSEHTQPAAACHRSYCS-----RSCLSG--------------------
MAV_0496|M.avium_104 LYNEHLAFPLNRPGFDGGSCYWISTSGWSVRFVA----------------
Mvan_0416|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_4200|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_4550|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_3782|M.marinum_M --------------------------------------------------
TH_3017|M.thermoresistible__bu --------------------------------------------------
Mb3360|M.bovis_AF2122/97 ALYRIAKRRFGEVPEPFTVTAHHRRLLIANVVHEALLQRASRKLPPSVRE
Rv0797|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0496|M.avium_104 --------------------------------------------------
Mvan_0416|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_4200|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_4550|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_3782|M.marinum_M --------------------------------------------------
TH_3017|M.thermoresistible__bu --------------------------------------------------
Mb3360|M.bovis_AF2122/97 LAVFWTARSIGCSWCVDFGAMLQRLDGLDVDRLTDIDNYATSSKFSDDER
Rv0797|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0496|M.avium_104 --------------------------------------------------
Mvan_0416|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_4200|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_4550|M.smegmatis_MC2_155 --------------------------------------------------
MMAR_3782|M.marinum_M --------------------------------------------------
TH_3017|M.thermoresistible__bu --------------------------------------------------
Mb3360|M.bovis_AF2122/97 AAIAYAEAMTADPHSVTDEQVADLRARFGEAGVIELTYQIGVENMRARMN
Rv0797|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0496|M.avium_104 --------------------------------------------------
Mvan_0416|M.vanbaalenii_PYR-1 -------------------------------
Mflv_4200|M.gilvum_PYR-GCK -------------------------------
MSMEG_4550|M.smegmatis_MC2_155 -------------------------------
MMAR_3782|M.marinum_M -------------------------------
TH_3017|M.thermoresistible__bu -------------------------------
Mb3360|M.bovis_AF2122/97 SALGITEQGFNSGDACRVPWAAPDVPSAESR
Rv0797|M.tuberculosis_H37Rv -------------------------------
MAV_0496|M.avium_104 -------------------------------