For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TTRSSELVPSDMKGLWGFVPACSTPDAADVNAVDTIDTDALASLVDRLVRDGVDGIVTTGSAGESHTLSD DEYRTLITTVVETVNARVPVFVGASTLNTRDSIRRARVIADLGADGIMSGPPMYLPQTAENAVQYYKDLA EAVPELAIMIYQNPHAFRITLPPGAFRELAQIRNIVALKQTSMDIFNVIGAIKAVKDKMSVLVLDQLMYP AMMFGAAGAWSIDVCMGPWPALSLRDACQRGDWTEAATIADQMQAPFRTLGLTMEEFQAMQSAWWKIAID TAGYGRAGAARPPFVHIPQTVVDSAHRYGERWAGLAERYHRSREAAGLPPAAANVAAASS*
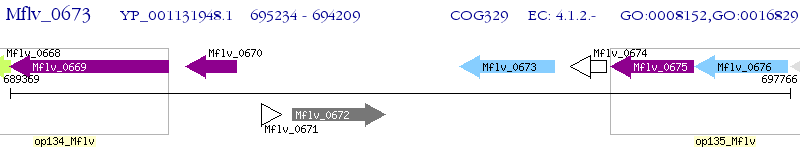
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0673 | - | - | 100% (341) | dihydrodipicolinate synthetase |
| M. gilvum PYR-GCK | Mflv_0585 | - | 0.0 | 100.00% (341) | dihydrodipicolinate synthetase |
| M. gilvum PYR-GCK | Mflv_0676 | - | 4e-54 | 35.29% (323) | dihydrodipicolinate synthetase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2774c | dapA | 4e-19 | 31.74% (230) | dihydrodipicolinate synthase |
| M. tuberculosis H37Rv | Rv2753c | dapA | 4e-19 | 31.74% (230) | dihydrodipicolinate synthase |
| M. leprae Br4923 | MLBr_01513 | dapA | 1e-20 | 32.17% (230) | dihydrodipicolinate synthase |
| M. abscessus ATCC 19977 | MAB_1047c | - | 8e-27 | 34.70% (219) | dihydrodipicolinate synthetase DapA |
| M. marinum M | MMAR_2154 | dapA_1 | 6e-23 | 32.30% (226) | dihydrodipicolinate synthase DapA_1 |
| M. avium 104 | MAV_3450 | - | 3e-24 | 31.86% (226) | dihydrodipicolinate synthetase family protein |
| M. smegmatis MC2 155 | MSMEG_2684 | dapA | 7e-21 | 30.22% (268) | dihydrodipicolinate synthase |
| M. thermoresistible (build 8) | TH_0834 | dapA | 6e-24 | 31.50% (254) | PROBABLE DIHYDRODIPICOLINATE SYNTHASE DAPA (DHDPS) |
| M. ulcerans Agy99 | MUL_1739 | dapA_1 | 4e-23 | 32.30% (226) | dihydrodipicolinate synthase DapA_1 |
| M. vanbaalenii PYR-1 | Mvan_0472 | - | 0.0 | 99.12% (341) | dihydrodipicolinate synthetase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb2774c|M.bovis_AF2122/97 ----MTTVGFDVAARLGTLLTAMVTP----FSGDGSLDTATAARLANHLV
Rv2753c|M.tuberculosis_H37Rv ----MTTVGFDVAARLGTLLTAMVTP----FSGDGSLDTATAARLANHLV
MLBr_01513|M.leprae_Br4923 ----MTTVGFDVPARLGTLLTAMVTP----FDADGSVDTAAATRLANRLV
MSMEG_2684|M.smegmatis_MC2_155 ----MSTSGIDVTAQLGTLLTAMVTP----FKPDGSLDLDAAAQLAARLV
TH_0834|M.thermoresistible__bu ----VSTGGFDVTAQLGTVLTAMVTP----FKKDGSLDLDAAASLANRLV
MMAR_2154|M.marinum_M -------MAEPGAPTIHGIIAYPVTP----FTAEGDVDARRLAALVDRLV
MUL_1739|M.ulcerans_Agy99 -------MAEPGAPTIHGIIAYPVTP----FTAEGDVDARRLAALVDRLV
MAV_3450|M.avium_104 -------MTEP---KIHGIIAYPVTP----FADNG-IDTQRLAALVERLV
MAB_1047c|M.abscessus_ATCC_199 -------MTST--PQFHGIIAYPVTP----FLPDDTVDIDRLAELVSRLV
Mflv_0673|M.gilvum_PYR-GCK MTTRSSELVPSDMKGLWGFVPACSTPDAADVNAVDTIDTDALASLVDRLV
Mvan_0472|M.vanbaalenii_PYR-1 MTTRSSELVPSDMKGLWGFVPACSTPDAADVNAVDTIDTDALASLVDRLV
: .:. ** . . :* : *. :**
Mb2774c|M.bovis_AF2122/97 DQGCDGLVVSGTTGESPTTTDGEKIELLRAVLEAVGDRARVIAGAGTYDT
Rv2753c|M.tuberculosis_H37Rv DQGCDGLVVSGTTGESPTTTDGEKIELLRAVLEAVGDRARVIAGAGTYDT
MLBr_01513|M.leprae_Br4923 DAGCDGLVLSGTTGESPTTTDDEKLQLLRVVLEAVGDRARVIAGAGSYDT
MSMEG_2684|M.smegmatis_MC2_155 DSGCDGLVLSGTTGESPTTSDDEKIALLRTVVDAVGDRARVIAGAGSYDT
TH_0834|M.thermoresistible__bu DAGCDGLVVSGTTGEAPTTTDDEKLQLLRTVLDAVGDRARIIAGAGTYDT
MMAR_2154|M.marinum_M CSGVHAIAALGSTGELAYLDEAEFDTAVDTAVAAVSGRLPVVVGVSDVTT
MUL_1739|M.ulcerans_Agy99 CSGVHAIAALGSTGELAYLDEAEFDTAVDTAVAAVSGRLPVVVGVSDVTT
MAV_3450|M.avium_104 SAGVHAIAPLGSTGELAYLEEPEVDAVIDTTLSAVAGRVPVIVGVSDVTT
MAB_1047c|M.abscessus_ATCC_199 DDGAHAIAPLGSTGESAYLEEREFDAVVDTTVAAVNKRVPVIVGASDLTT
Mflv_0673|M.gilvum_PYR-GCK RDGVDGIVTTGSAGESHTLSDDEYRTLITTVVETVNARVPVFVGASTLNT
Mvan_0472|M.vanbaalenii_PYR-1 RDGVDGIVTTGSAGESHTLSDDEYRTLITTVVETVNARVPVFVGASTLNT
* ..:. *::** : * : ..: :* * :..*.. *
Mb2774c|M.bovis_AF2122/97 AHSIRLAKACAAEGAHGLLVVTPYYSKPPQRGLQAHFTAVADAT-ELPML
Rv2753c|M.tuberculosis_H37Rv AHSIRLAKACAAEGAHGLLVVTPYYSKPPQRGLQAHFTAVADAT-ELPML
MLBr_01513|M.leprae_Br4923 AHSVRLVKACAGEGAHGLLVVTPYYSKPPQTGLFAHFTAVADAT-ELPVL
MSMEG_2684|M.smegmatis_MC2_155 AHSVHLAKASAEAGAHGLLVVTPYYSRPAQAGLLAHFTAVADAT-DLPVV
TH_0834|M.thermoresistible__bu AHSVRLAKACQDAGAHGLLLVTPYYSRPPQDGLYAHFTTIADAV-EVPIV
MMAR_2154|M.marinum_M AKTIRRARYAQHVGADAVMILPVSYWKLNEHEIFQHYRSVGDAI-TIPIM
MUL_1739|M.ulcerans_Agy99 AKTIRRARYAQHVGADAVMILPVSYWKLNEHEIFQHYRSVGDAI-TIPIM
MAV_3450|M.avium_104 AKTIRRAQYAQRAGADAVMILPVSYWKLTEREIFHHYRSISDAI-SIPIM
MAB_1047c|M.abscessus_ATCC_199 ANTIRRARHAQQAGADAVMVLPISYWKLSDREIAQHYASVAAAI-DIPLM
Mflv_0673|M.gilvum_PYR-GCK RDSIRRARVIADLGADGIMSGPPMYLPQTAENAVQYYKDLAEAVPELAIM
Mvan_0472|M.vanbaalenii_PYR-1 RDSIRRARVIADLGADGIMSGPPMYLPQTAENAVQYYKDLAEAVPELAIM
.::: .: **..:: . * :: :. * :.::
Mb2774c|M.bovis_AF2122/97 LYDIPGRSAVPIEPDTIR-ALASHPNIVGVKDAKADLHSGAQIMADTG--
Rv2753c|M.tuberculosis_H37Rv LYDIPGRSAVPIEPDTIR-ALASHPNIVGVKDAKADLHSGAQIMADTG--
MLBr_01513|M.leprae_Br4923 LYDIPGRSVVPIEPDTIR-ALASHPNIVGVKEAKADLYSGARIMADTG--
MSMEG_2684|M.smegmatis_MC2_155 LYDIPPRSVVPIAWDTLR-KLAEHPNIVAVKDAKGDLPGGGQIIAETG--
TH_0834|M.thermoresistible__bu LYDIPPRSVVPLAWDTIR-RLSEHRNIVAIKDAKGDLHGAAQIIAETG--
MMAR_2154|M.marinum_M AYNNPATSGVDMRPELLVRMFESIDNLTMVKESTGDLSRMRRIAELSGGR
MUL_1739|M.ulcerans_Agy99 AYNNPATSGVDMRPELLVRMFESIDNLTMVKESTGDLSRMRRIAELSGGR
MAV_3450|M.avium_104 AYNNPATSGVDMPPELLVRMFESIDNVAMVKESTGDLTRMRRIAELSGNR
MAB_1047c|M.abscessus_ATCC_199 AYNNPATSGVDMKPELLVSMFGDIDNFTMVKESTGDLNRMLEIKRLTNGL
Mflv_0673|M.gilvum_PYR-GCK IYQNPHAFRITLPPGAFR-ELAQIRNIVALKQTSMDIFNVIGAIKAVKDK
Mvan_0472|M.vanbaalenii_PYR-1 IYQNPHAFRITLPPGAFR-ELAQIRNIVALKQTSMDIFNVIGAIKAVKEK
*: * : : : : . *.. :*::. *:
Mb2774c|M.bovis_AF2122/97 LAYYSGDDALNLPWLAMGATGFISVIAHLAAGQLRELLSAFGSGDIATAR
Rv2753c|M.tuberculosis_H37Rv LAYYSGDDALNLPWLAMGATGFISVIAHLAAGQLRELLSAFGSGDIATAR
MLBr_01513|M.leprae_Br4923 LAYYSGDDALNLPWLAVGAIGFISVISHLAAGQLRELLSAFGSGDITTAR
MSMEG_2684|M.smegmatis_MC2_155 LAYYSGDDALNLPWLAMGALGFISVWGHLAAGQLRDMLTAFNSGDVATAR
TH_0834|M.thermoresistible__bu LAYYSGDDSLNLPWLAVGAVGFISVWGHLAAGQLRDMLTAFNSGDMTTAR
MMAR_2154|M.marinum_M LPFYNGSNPLVLDALKVGAAGWCTAAPNLWPQPCIDLYEAVCAGDLEKAQ
MUL_1739|M.ulcerans_Agy99 LPFYNGSNPLVLDALKVGAAGWCTAAPNLWPQPCIDLYEAVCAGDLEKAQ
MAV_3450|M.avium_104 LPFYNGSNPLVLDALKAGASGWCTAAPCLRPQPCIDLYDAVRANDLDTAQ
MAB_1047c|M.abscessus_ATCC_199 LPFYNGSNPLVLDALNAGASGWCTAAPCLAPQPCLDLYEAVRAGRDDDAA
Mflv_0673|M.gilvum_PYR-GCK MSVLVLD-QLMYPAMMFGAAGAWSIDVCMGPWPALSLRDACQRGDWTEAA
Mvan_0472|M.vanbaalenii_PYR-1 MSVLVLD-QLMYPAMMFGAAGAWSIDVCMGPWPALSLRDACQRGDWTEAA
:. . * : ** * : : . .: * . *
Mb2774c|M.bovis_AF2122/97 KINIAVAP------LCNAMSRLGGVTLSKAGLRLQGI-DVGDPRLPQVAA
Rv2753c|M.tuberculosis_H37Rv KINIAVAP------LCNAMSRLGGVTLSKAGLRLQGI-DVGDPRLPQVAA
MLBr_01513|M.leprae_Br4923 KINVAIGP------LCSAMDRLGGVTMSKAGLRLQGI-DVGDPRLPQMPA
MSMEG_2684|M.smegmatis_MC2_155 KINVNLAP------LAAAQARLGGVTMSKAGLRLQGF-DAGEPRLPQIPA
TH_0834|M.thermoresistible__bu KIAVTLSP------LNAAQTRLGGVSMSKAGLRLLGI-EVGDPRLPQVPP
MMAR_2154|M.marinum_M ILYDDLKP------LLEFIVAGGLATTVKAGLGLLGF-PVGDPRPPLLPL
MUL_1739|M.ulcerans_Agy99 ILYDDLKP------LLEFIVAGRLATTVKAGLGLLGF-PVGDPRPPLLPL
MAV_3450|M.avium_104 RLYDDLKP------LLTFIVAGGLATTVKAGLELLDF-KVGDPRAPLLPL
MAB_1047c|M.abscessus_ATCC_199 SLYTALKP------LLQFIVAGGLPTTVKAGLELQGR-PVGDPRRPLLPL
Mflv_0673|M.gilvum_PYR-GCK TIADQMQAPFRTLGLTMEEFQAMQSAWWKIAIDTAGYGRAGAARPPFVHI
Mvan_0472|M.vanbaalenii_PYR-1 AIADQMQAPFRTLGLTMEEFQAMQSAWWKMAIDTAGYGRAGAARPPFVHI
: : . * : * .: . .* .* * :
Mb2774c|M.bovis_AF2122/97 --------------------------TPEQIDALAADMRAASVLR
Rv2753c|M.tuberculosis_H37Rv --------------------------TPEQIDALAADMRAASVLR
MLBr_01513|M.leprae_Br4923 --------------------------TAEQIDELAVDMRAASVLR
MSMEG_2684|M.smegmatis_MC2_155 --------------------------TTEEVEALAADMRAATVLR
TH_0834|M.thermoresistible__bu --------------------------GPEQLRLLADDMRAAAVLR
MMAR_2154|M.marinum_M --------------------------DEHGRAKLGRLLAGC----
MUL_1739|M.ulcerans_Agy99 --------------------------DEHGRAKLGRLLAGC----
MAV_3450|M.avium_104 --------------------------DEQGRAELRKLLAA-----
MAB_1047c|M.abscessus_ATCC_199 --------------------------DPEGRETLKGILASTP---
Mflv_0673|M.gilvum_PYR-GCK PQTVVDSAHRYGERWAGLAERYHRSREAAGLPPAAANVAAASS--
Mvan_0472|M.vanbaalenii_PYR-1 PQTVVDSAHRYGERWAGLAERYHRSREAAGLPPAAANVAAASS--
: .