For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TDVASRELGLGFLGIGQAVARIFQQYPDMSTLPYRVVAAADTRSHSLARFATEFSGQTYTNAEQLCADPS VDVVYIATPPELHREHALMAARHGKHMIVEKPLAMSIDDCTAMVEAAQAAGVQLMAGHTHSFDAPVRAMA ELVASGELGELLMVNTWNFNDFNRRPWPTSELRSTSGPVLNQGPHQVDIVRQIVGGVARTVRASTIWDDV RECVGGYTCHLGFESGASATLVYDARAFFDVAELHSWVAEDGGRRSPQANQRVANNFAALSQNPDQLEVA LEAQKEQGRYGAATTTEESQELWGYSAPGEIIHHPYFGLTVVSCERGAIRQSPDGLVVYGQNGLQEIPVA RTMRGRAAELAELHRAITEDRPVQHDGRWGRATLEVCFAILQSAYEDREVVLSHQVSL*
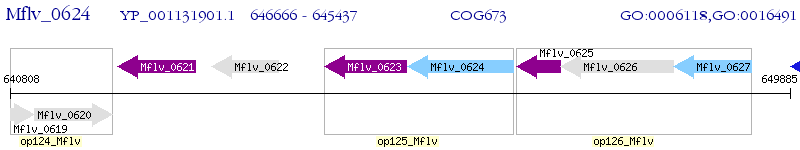
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0624 | - | - | 100% (409) | oxidoreductase domain-containing protein |
| M. gilvum PYR-GCK | Mflv_0542 | - | 0.0 | 100.00% (409) | oxidoreductase domain-containing protein |
| M. gilvum PYR-GCK | Mflv_2611 | - | 3e-10 | 28.41% (176) | oxidoreductase domain-containing protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | - | - | - | - | - |
| M. tuberculosis H37Rv | - | - | - | - | - |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2306c | - | 7e-15 | 28.14% (231) | putative oxidoreductase |
| M. marinum M | MMAR_2628 | - | 1e-10 | 26.77% (198) | hypothetical protein MMAR_2628 |
| M. avium 104 | MAV_1941 | - | 4e-11 | 27.07% (314) | oxidoreductase family protein, NAD-binding Rossmann fold |
| M. smegmatis MC2 155 | MSMEG_4650 | - | 4e-11 | 26.02% (269) | oxidoreductase YisS |
| M. thermoresistible (build 8) | TH_3104 | - | 1e-05 | 26.62% (139) | PUTATIVE oxidoreductase domain protein |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0541 | - | 0.0 | 100.00% (409) | oxidoreductase domain-containing protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0624|M.gilvum_PYR-GCK MTDVASRELGLGFLGIGQAVARIFQQYPDMSTLPYRVVAAADTRSHSLAR
Mvan_0541|M.vanbaalenii_PYR-1 MTDVASRELGLGFLGIGQAVARIFQQYPDMSTLPYRVVAAADTRSHSLAR
MSMEG_4650|M.smegmatis_MC2_155 -------MTTLGVIGLGRIGAFHVDTLSSLDGVDG--LVVADERPDAVAA
MAV_1941|M.avium_104 ------MTLRVGIIGVGWGAHVQVPGFRAAKGYEP--VALCARTPERLER
TH_3104|M.thermoresistible__bu --------MRVLVIGTGFGRQAMAPVYRGLG------FDVEVVSPRDESA
MAB_2306c|M.abscessus_ATCC_199 ----MPESIRWGIVGPGRIAGNVARDFPLTPRAEL--AAVASRSLPRAQD
MMAR_2628|M.marinum_M -MTGHGSPLRIGILGAARIAP-QALVKPAQGNVEAVVAAVAARDGARAQA
.:* .
Mflv_0624|M.gilvum_PYR-GCK FATEFS-GQTYTNAEQLCADPSVDVVYIATPPELHREHALMAARHGKHMI
Mvan_0541|M.vanbaalenii_PYR-1 FATEFS-GQTYTNAEQLCADPSVDVVYIATPPELHREHALMAARHGKHMI
MSMEG_4650|M.smegmatis_MC2_155 VAAKHG-AKPADSVEELLDS-GVDGVVVAAATPAHAELTLAAVHRGLPTF
MAV_1941|M.avium_104 VADKLGIEETSTDWESFVTRDDLDVISVATPTVLHRDMTLAALAAGKPVL
TH_3104|M.thermoresistible__bu LHRALN--------------AGVDLVSVHSPPFLHRQHVTAAVDRGYPVL
MAB_2306c|M.abscessus_ATCC_199 FAGAHQIPRAYGSYRELLADPDIDAVYIATPHPQHRQTAIAALTAGKAIL
MMAR_2628|M.marinum_M FAAKHGISRVHSSYEALIADPTLDAVYIPLPNGLHGRWTQAALTAGKHVL
. :* : : . * . * * :
Mflv_0624|M.gilvum_PYR-GCK VEKPLAMSIDDCTAMVEAAQAAGVQLMAGHTHSFDAPVRAMAELVASGEL
Mvan_0541|M.vanbaalenii_PYR-1 VEKPLAMSIDDCTAMVEAAQAAGVQLMAGHTHSFDAPVRAMAELVASGEL
MSMEG_4650|M.smegmatis_MC2_155 CEKPIAATAAESARVAEAIARSGVPVQVGYQRRFDAAFAAAKREVDSGSL
MAV_1941|M.avium_104 CEKPLAGDLDAARDMVRAAAEANVPTACCFENRWNPDWLAVADRVRSGFL
TH_3104|M.thermoresistible__bu CDKPFGRNADEARAMRDHAARRAVPTFLNFELRHQPARAEFKKLADSGVI
MAB_2306c|M.abscessus_ATCC_199 VEKTFTATVSGAQEVIDIARARGVFAMEAMWTRFQPAIVAAKDLVDDGAI
MMAR_2628|M.marinum_M CEKPFTANAAEADEIAQLAATTDRVVMEAFHYRYHPLTLRIEQILASGEL
:*.: . : .. .* :
Mflv_0624|M.gilvum_PYR-GCK G-ELLMVNTWNFNDFNRRPWPTS-----ELRSTSGPVLNQGPHQVDIVRQ
Mvan_0541|M.vanbaalenii_PYR-1 G-ELLMVNTWNFNDFNRRPWPTS-----ELRSTSGPVLNQGPHQVDIVRQ
MSMEG_4650|M.smegmatis_MC2_155 G-ALHTVRST---TMDPAPPSMD-----YIKGSGGIFRDCAVHDFDALRW
MAV_1941|M.avium_104 G-TQYLARVSRSASYWHPSHPLQAHWMYDREQGGGYLAGMLVHDLDFLCS
TH_3104|M.thermoresistible__bu GTPRHLSWTFFGAGLRGRPHGWIN----ERDTGGGWIGANGAHLIDYTRY
MAB_2306c|M.abscessus_ATCC_199 GEVRQVQADLGVDRPFDREDRLFN-----PALGGGALLDLGVYVVSLAQY
MMAR_2628|M.marinum_M GTLQRVETSMCFPLPKFSDIRYN------YALAGGATMDAGCYAVHMART
* .* : .
Mflv_0624|M.gilvum_PYR-GCK IVGGVARTVRASTIWDDVRECVGGYTCHLGFESGASATLVYDARAFFDVA
Mvan_0541|M.vanbaalenii_PYR-1 IVGGVARTVRASTIWDDVRECVGGYTCHLGFESGASATLVYDARAFFDVA
MSMEG_4650|M.smegmatis_MC2_155 ITG-------------------------------QNAVEVYATGTVQGDP
MAV_1941|M.avium_104 LLG----------------------------RPVSVCAEVRTSDPVRELP
TH_3104|M.thermoresistible__bu LFG---------------------------SEITGCGGVARIEDPHRPDP
MAB_2306c|M.abscessus_ATCC_199 FLG----------------------------------------DPAAVVA
MMAR_2628|M.marinum_M FGG---------------------------------------GTPEVVSA
: * . .
Mflv_0624|M.gilvum_PYR-GCK E--LHSWVAEDGGRRSPQANQRVANNFAALSQNPDQLEVALEAQKEQGRY
Mvan_0541|M.vanbaalenii_PYR-1 E--LHSWVAEDGGRRSPQANQRVANNFAALSQNPDQLEVALEAQKEQGRY
MSMEG_4650|M.smegmatis_MC2_155 R--FTEYGDVDTAAVVVRFDGGALGIVSAARYNARGYDCRLEVHGFDN--
MAV_1941|M.avium_104 DGGTLHVTADDTAAVLMRMESGVTAVLSVSVMGAHADHYRLELFGSDG--
TH_3104|M.thermoresistible__bu QGRPRRATAEDAYSAWFVTENGCTATHDTAFAAAVALPPRMVLTGSAGAL
MAB_2306c|M.abscessus_ATCC_199 HGSLFPTGVDAEAGLLLRYDDGRTATLLCSLLHHT--PGQARIFGTEGWI
MMAR_2628|M.marinum_M QAKRRDSEVDRAMTAELRFPAGHSGRIQCSMWSSHLLNMSARVVGDRGEL
.
Mflv_0624|M.gilvum_PYR-GCK GAATTTEESQELWGYSAPGEIIHHPYFGLTVVSCERGAIRQSPDGLVVYG
Mvan_0541|M.vanbaalenii_PYR-1 GAATTTEESQELWGYSAPGEIIHHPYFGLTVVSCERGAIRQSPDGLVVYG
MSMEG_4650|M.smegmatis_MC2_155 SIVAGWDQGTPLQNMDPRNDFPTGPYHHFFMDRFT-EAFRSELAGFVEVV
MAV_1941|M.avium_104 TIIGDGDLRSAAYSLGAATDDGLRSLAVDEREPAHPDRLPRGLAGHASRA
TH_3104|M.thermoresistible__bu ELIGD----SRLIVHRADADPMTHEFPPAPPRSFYEPALTPWLEQVADAL
MAB_2306c|M.abscessus_ATCC_199 DILPRFHHPREIVLHRKG------AAPEPMVRPPVGGGYSHELAEVTECL
MMAR_2628|M.marinum_M RVLNPVVPQLFHRLTVQS------AIGRRVERFPRRASYAYQLDAFTGAI
.
Mflv_0624|M.gilvum_PYR-GCK QNGLQEIPVARTMRGRAAELAELHRAITEDRPVQHDGRWGRATLEVCFAI
Mvan_0541|M.vanbaalenii_PYR-1 QNGLQEIPVARTMRGRAAELAELHRAITEDRPVQHDGRWGRATLEVCFAI
MSMEG_4650|M.smegmatis_MC2_155 KG----NPIPGATVADAVEVAWMAEAATES-------------LRRGAPV
MAV_1941|M.avium_104 MALMLQDWLP-AFDGAPSNVATFEDGLLSL-------------AIIDAAR
TH_3104|M.thermoresistible__bu RTG----RPATPSFDDGVAVAEVMDRLRAN--------------VIEQVM
MAB_2306c|M.abscessus_ATCC_199 LNG--RAQSSVMPLADTLAVQRVLNTACEQ-------------LGVDHTE
MMAR_2628|M.marinum_M LRG----EAVKTTPQDAVQNMTVIDAIYRA---------------AGLPL
.
Mflv_0624|M.gilvum_PYR-GCK LQSAYEDREVVLSHQVSL
Mvan_0541|M.vanbaalenii_PYR-1 LQSAYEDREVVLSHQVSL
MSMEG_4650|M.smegmatis_MC2_155 RIEEVRNR----------
MAV_1941|M.avium_104 RSAEGGGWEPVQA-----
TH_3104|M.thermoresistible__bu Q-----------------
MAB_2306c|M.abscessus_ATCC_199 DPADLD------------
MMAR_2628|M.marinum_M RNPS--------------