For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
RDTKLTVDDITGVVGIIPTPSIPTADQPGTAFSVDLDEAAKLADAMVRGGVDVLMTTGTFGECASLTWDE LQSFVATVVDAVAGRIPVFAGATTLNTRDTIARGRRLGELGADGLFVGRPMWLPLDDAGIVRFYRDVAEA VPNMALVVYDNPGAFKGKIGTPAYEALSQIPQVVAAKHLGLLSGSAFLSDLRAVGGRVRLLPLETDWYYF ARLFPEEVTACWSGNVACGPAPVTHLRDLIRARRWDDCQALTDELEGALETLYPGGNFAEFLKYSIQIDN AQFQAAGFMRTGPTRPPYTEVPESYLAGGREAGKNWAALQQRYASLAVSNH*
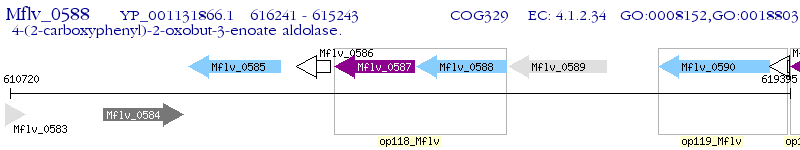
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0588 | - | - | 100% (332) | dihydrodipicolinate synthetase |
| M. gilvum PYR-GCK | Mflv_0676 | - | 0.0 | 100.00% (332) | dihydrodipicolinate synthetase |
| M. gilvum PYR-GCK | Mflv_0673 | - | 4e-54 | 35.29% (323) | dihydrodipicolinate synthetase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2774c | dapA | 1e-13 | 29.89% (184) | dihydrodipicolinate synthase |
| M. tuberculosis H37Rv | Rv2753c | dapA | 2e-13 | 29.89% (184) | dihydrodipicolinate synthase |
| M. leprae Br4923 | MLBr_01513 | dapA | 6e-15 | 29.89% (184) | dihydrodipicolinate synthase |
| M. abscessus ATCC 19977 | MAB_1047c | - | 4e-15 | 28.08% (260) | dihydrodipicolinate synthetase DapA |
| M. marinum M | MMAR_1962 | dapA | 1e-15 | 32.32% (164) | dihydrodipicolinate synthase DapA |
| M. avium 104 | MAV_3450 | - | 5e-16 | 26.46% (257) | dihydrodipicolinate synthetase family protein |
| M. smegmatis MC2 155 | MSMEG_2684 | dapA | 4e-17 | 33.73% (169) | dihydrodipicolinate synthase |
| M. thermoresistible (build 8) | TH_0834 | dapA | 1e-16 | 31.84% (179) | PROBABLE DIHYDRODIPICOLINATE SYNTHASE DAPA (DHDPS) |
| M. ulcerans Agy99 | MUL_2184 | dapA | 9e-16 | 32.32% (164) | dihydrodipicolinate synthase |
| M. vanbaalenii PYR-1 | Mvan_0469 | - | 0.0 | 99.70% (332) | dihydrodipicolinate synthetase |
CLUSTAL 2.0.9 multiple sequence alignment
MSMEG_2684|M.smegmatis_MC2_155 --MSTSGIDVTAQLGTLLTAMVTPFKP----DGSLDLDAAAQLAARLVDS
TH_0834|M.thermoresistible__bu --VSTGGFDVTAQLGTVLTAMVTPFKK----DGSLDLDAAASLANRLVDA
Mb2774c|M.bovis_AF2122/97 --MTTVGFDVAARLGTLLTAMVTPFSG----DGSLDTATAARLANHLVDQ
Rv2753c|M.tuberculosis_H37Rv --MTTVGFDVAARLGTLLTAMVTPFSG----DGSLDTATAARLANHLVDQ
MMAR_1962|M.marinum_M --MSTVGFDVPARLGTLLTAMVTPFGS----DGALDLAAAARLANHLVDQ
MUL_2184|M.ulcerans_Agy99 --MSTVGFDVPARLGTLLTAMVTPFGS----DGALDLAAAARLANHLVDQ
MLBr_01513|M.leprae_Br4923 --MTTVGFDVPARLGTLLTAMVTPFDA----DGSVDTAAATRLANRLVDA
MAB_1047c|M.abscessus_ATCC_199 -------MTSTPQFHGIIAYPVTPFLP----DDTVDIDRLAELVSRLVDD
MAV_3450|M.avium_104 --------MTEPKIHGIIAYPVTPFA-----DNGIDTQRLAALVERLVSA
Mflv_0588|M.gilvum_PYR-GCK MRDTKLTVDDITGVVGIIPTPSIPTADQPGTAFSVDLDEAAKLADAMVRG
Mvan_0469|M.vanbaalenii_PYR-1 MRDTKLTVDDITGVVGIIPTPSIPTADQPGTAFSVDLDEAAKLADAMVRG
. . ::. * :* : *. :*
MSMEG_2684|M.smegmatis_MC2_155 GCDGLVLSGTTGESPTTSDDEKIALLRTVVDAVGDRARVIAGAGSYDTAH
TH_0834|M.thermoresistible__bu GCDGLVVSGTTGEAPTTTDDEKLQLLRTVLDAVGDRARIIAGAGTYDTAH
Mb2774c|M.bovis_AF2122/97 GCDGLVVSGTTGESPTTTDGEKIELLRAVLEAVGDRARVIAGAGTYDTAH
Rv2753c|M.tuberculosis_H37Rv GCDGLVVSGTTGESPTTTDGEKIELLRAVLEAVGDRARVIAGAGTYDTAH
MMAR_1962|M.marinum_M GCDGLVVSGTTGESPTTTDDEKRELLRVVLEAVGDRARVIAGAGTNDTAH
MUL_2184|M.ulcerans_Agy99 GCDGLVVSGTTGESPTTTDDEKRELLRVVLEAVGDRARVIAGAGTNDTAH
MLBr_01513|M.leprae_Br4923 GCDGLVLSGTTGESPTTTDDEKLQLLRVVLEAVGDRARVIAGAGSYDTAH
MAB_1047c|M.abscessus_ATCC_199 GAHAIAPLGSTGESAYLEEREFDAVVDTTVAAVNKRVPVIVGASDLTTAN
MAV_3450|M.avium_104 GVHAIAPLGSTGELAYLEEPEVDAVIDTTLSAVAGRVPVIVGVSDVTTAK
Mflv_0588|M.gilvum_PYR-GCK GVDVLMTTGTFGECASLTWDELQSFVATVVDAVAGRIPVFAGATTLNTRD
Mvan_0469|M.vanbaalenii_PYR-1 GVDVLMTTGTFGECASLTWDELQSFVATVVDAVAGRIPVFAGATTLNTRD
* . : *: ** . * .: ..: ** * ::.*. * .
MSMEG_2684|M.smegmatis_MC2_155 SVHLAKASAEAGAHGLLVVTPYYSRPAQAGLLAHFTAVADAT-DLPVVLY
TH_0834|M.thermoresistible__bu SVRLAKACQDAGAHGLLLVTPYYSRPPQDGLYAHFTTIADAV-EVPIVLY
Mb2774c|M.bovis_AF2122/97 SIRLAKACAAEGAHGLLVVTPYYSKPPQRGLQAHFTAVADAT-ELPMLLY
Rv2753c|M.tuberculosis_H37Rv SIRLAKACAAEGAHGLLVVTPYYSKPPQRGLQAHFTAVADAT-ELPMLLY
MMAR_1962|M.marinum_M SIGLAKAAAAEGAHGLLVVTPYYSKPPQSGLLAHFTAVADAT-ELPVLLY
MUL_2184|M.ulcerans_Agy99 SIGLAKAAAAEGAHGLLVVTPYYSKPPQSGLLAHFTAVADAT-ELPVLLY
MLBr_01513|M.leprae_Br4923 SVRLVKACAGEGAHGLLVVTPYYSKPPQTGLFAHFTAVADAT-ELPVLLY
MAB_1047c|M.abscessus_ATCC_199 TIRRARHAQQAGADAVMVLPISYWKLSDREIAQHYASVAAAI-DIPLMAY
MAV_3450|M.avium_104 TIRRAQYAQRAGADAVMILPVSYWKLTEREIFHHYRSISDAI-SIPIMAY
Mflv_0588|M.gilvum_PYR-GCK TIARGRRLGELGADGLFVGRPMWLPLDDAGIVRFYRDVAEAVPNMALVVY
Mvan_0469|M.vanbaalenii_PYR-1 TIARGRRLGELGADGLFVGRPMWLPLDDAGIVRFYRDVAEAVPNMALVVY
:: : **..::: : : : .: :: * .:.:: *
MSMEG_2684|M.smegmatis_MC2_155 DIPPRSVVPIAWDTLR-KLAEHPNIVAVKDA--KGDLPGGGQIIAETG--
TH_0834|M.thermoresistible__bu DIPPRSVVPLAWDTIR-RLSEHRNIVAIKDA--KGDLHGAAQIIAETG--
Mb2774c|M.bovis_AF2122/97 DIPGRSAVPIEPDTIR-ALASHPNIVGVKDA--KADLHSGAQIMADTG--
Rv2753c|M.tuberculosis_H37Rv DIPGRSAVPIEPDTIR-ALASHPNIVGVKDA--KADLHSGAQIMADTG--
MMAR_1962|M.marinum_M DIPGRSAVPIESETIR-ALAKHPNIVGVKDA--KADLPSGAQIMADTG--
MUL_2184|M.ulcerans_Agy99 DIPGRSAVPIESETIR-ALAKHPNIVGVKDA--KADLPSGAQIMADTG--
MLBr_01513|M.leprae_Br4923 DIPGRSVVPIEPDTIR-ALASHPNIVGVKEA--KADLYSGARIMADTG--
MAB_1047c|M.abscessus_ATCC_199 NNPATSGVDMKPELLVSMFGDIDNFTMVKES--TGDLNRMLEIKRLTNGL
MAV_3450|M.avium_104 NNPATSGVDMPPELLVRMFESIDNVAMVKES--TGDLTRMRRIAELSGNR
Mflv_0588|M.gilvum_PYR-GCK DNPGAFKGKIGTPAYE-ALSQIPQVVAAKHLGLLSGSAFLSDLRAVGGRV
Mvan_0469|M.vanbaalenii_PYR-1 DNPGAFKGKIGTPAYE-ALSQIPQVVAAKHLGLLSGSAFLSDLRAVSGRV
: * : : . :.. *. .. : .
MSMEG_2684|M.smegmatis_MC2_155 -LAYYSGDDALNLPWLAMGALGFISVWGHLAAGQLRDMLTAFNSGDVATA
TH_0834|M.thermoresistible__bu -LAYYSGDDSLNLPWLAVGAVGFISVWGHLAAGQLRDMLTAFNSGDMTTA
Mb2774c|M.bovis_AF2122/97 -LAYYSGDDALNLPWLAMGATGFISVIAHLAAGQLRELLSAFGSGDIATA
Rv2753c|M.tuberculosis_H37Rv -LAYYSGDDALNLPWLAMGATGFISVIAHLAAGQLRELLSAFGSGDIATA
MMAR_1962|M.marinum_M -LAYYSGDDALNLPWLAMGAIGFISVIGHLAAGQLREMLSAFSSGDVATA
MUL_2184|M.ulcerans_Agy99 -LAYYSGDDALNLPWLAMGAIGFISVIGHLAAGQLREMLSAFSSGDVATA
MLBr_01513|M.leprae_Br4923 -LAYYSGDDALNLPWLAVGAIGFISVISHLAAGQLRELLSAFGSGDITTA
MAB_1047c|M.abscessus_ATCC_199 -LPFYNGSNPLVLDALNAGASGWCTAAPCLAPQPCLDLYEAVRAGRDDDA
MAV_3450|M.avium_104 -LPFYNGSNPLVLDALKAGASGWCTAAPCLRPQPCIDLYDAVRANDLDTA
Mflv_0588|M.gilvum_PYR-GCK RLLPLETDWYYFARLFPEEVTACWSGNVACGPAPVTHLRDLIRARRWDDC
Mvan_0469|M.vanbaalenii_PYR-1 RLLPLETDWYYFARLFPEEVTACWSGNVACGPAPVTHLRDLIRARRWDDC
* . . : . . : . .: . : .
MSMEG_2684|M.smegmatis_MC2_155 RKINVNLAPLAAAQARLG--------GVTMSKAGLRLQGFDAGEPRLPQI
TH_0834|M.thermoresistible__bu RKIAVTLSPLNAAQTRLG--------GVSMSKAGLRLLGIEVGDPRLPQV
Mb2774c|M.bovis_AF2122/97 RKINIAVAPLCNAMSRLG--------GVTLSKAGLRLQGIDVGDPRLPQV
Rv2753c|M.tuberculosis_H37Rv RKINIAVAPLCNAMSRLG--------GVTLSKAGLRLQGIDVGDPRLPQV
MMAR_1962|M.marinum_M RKINVAVSPLCNAMTRLG--------GVTLSKAGLRLQGIEVGDPRLPQV
MUL_2184|M.ulcerans_Agy99 RKINVAVSPLCNAMTRLG--------GVTLSKAGLRLQGIEVGDPRLPQV
MLBr_01513|M.leprae_Br4923 RKINVAIGPLCSAMDRLG--------GVTMSKAGLRLQGIDVGDPRLPQM
MAB_1047c|M.abscessus_ATCC_199 ASLYTALKPLLQFIVAGG--------LPTTVKAGLELQGRPVGDPRRPLL
MAV_3450|M.avium_104 QRLYDDLKPLLTFIVAGG--------LATTVKAGLELLDFKVGDPRAPLL
Mflv_0588|M.gilvum_PYR-GCK QALTDELEGALETLYPGGNFAEFLKYSIQIDNAQFQAAGFMRTGPTRPPY
Mvan_0469|M.vanbaalenii_PYR-1 QALTDELEGALETLYPGGNFAEFLKYSIQIDNAQFQAAGFMRTGPTRPPY
: : * :* :. . * *
MSMEG_2684|M.smegmatis_MC2_155 PATTE-----------EVEALAADMRAATVLR-
TH_0834|M.thermoresistible__bu PPGPE-----------QLRLLADDMRAAAVLR-
Mb2774c|M.bovis_AF2122/97 AATPE-----------QIDALAADMRAASVLR-
Rv2753c|M.tuberculosis_H37Rv AATPE-----------QIDALAADMRAASVLR-
MMAR_1962|M.marinum_M AATAE-----------QIDVLAADMRAASVLR-
MUL_2184|M.ulcerans_Agy99 AATAE-----------QIDVLAADMRAASVLR-
MLBr_01513|M.leprae_Br4923 PATAE-----------QIDELAVDMRAASVLR-
MAB_1047c|M.abscessus_ATCC_199 PLDPE-----------GRETLKGILASTP----
MAV_3450|M.avium_104 PLDEQ-----------GRAELRKLLAA------
Mflv_0588|M.gilvum_PYR-GCK TEVPESYLAGGREAGKNWAALQQRYASLAVSNH
Mvan_0469|M.vanbaalenii_PYR-1 TEVPESYLAGGREAGKNWAALQQRYASLAVSNH
. : * :