For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VAATDLLAGRATLSRSLRLLSAFRFEQTDPDRFYGALASDTVAMVDDLWRATTGTSTRNLTVLDVGGGPG YFASAFTDAGLHYVGVEPDPGEMHAAAPRTHSRAGSFVRASGTALPFADGSVDICLSSNVAEHVAQPWRL GEEMLRVTRPGGLAVLSYTVWLGPFGGHEMGLSHYLGGYRAAERYTRRHGHRPKNDYGSSLFAVSAADGL RWARSTGALAAAFPRYHPRWAWGLTRVPGVREFLVSNLVLVLRPT
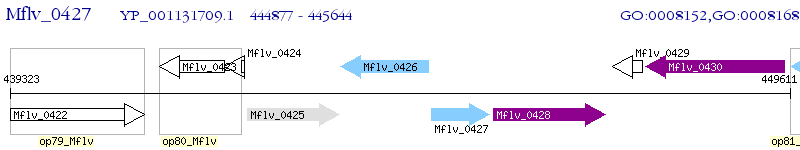
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0427 | - | - | 100% (255) | methyltransferase type 11 |
| M. gilvum PYR-GCK | Mflv_4260 | - | 5e-08 | 31.07% (103) | methyltransferase type 11 |
| M. gilvum PYR-GCK | Mflv_1100 | - | 2e-07 | 27.34% (128) | methyltransferase type 11 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0229c | - | 1e-108 | 74.02% (254) | methyltransferase (methylase) |
| M. tuberculosis H37Rv | Rv0224c | - | 1e-108 | 74.41% (254) | methyltransferase (methylase) |
| M. leprae Br4923 | MLBr_02584 | - | 1e-108 | 74.10% (251) | hypothetical protein MLBr_02584 |
| M. abscessus ATCC 19977 | MAB_4481 | - | 1e-107 | 72.55% (255) | hypothetical protein MAB_4481 |
| M. marinum M | MMAR_0473 | - | 1e-111 | 76.77% (254) | methyltransferase |
| M. avium 104 | MAV_4945 | - | 1e-111 | 76.10% (251) | SAM-dependent methyltransferase |
| M. smegmatis MC2 155 | MSMEG_6483 | - | 2e-08 | 26.79% (112) | methyltransferase type 11 |
| M. thermoresistible (build 8) | TH_2028 | - | 1e-113 | 77.25% (255) | POSSIBLE METHYLTRANSFERASE (METHYLASE) |
| M. ulcerans Agy99 | MUL_1123 | - | 1e-109 | 75.98% (254) | methyltransferase |
| M. vanbaalenii PYR-1 | Mvan_0241 | - | 1e-125 | 83.46% (254) | methyltransferase type 11 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0427|M.gilvum_PYR-GCK MAA--------------TDLLAGRATLSRSLRLLSAFRFEQTDPDRFYGA
Mvan_0241|M.vanbaalenii_PYR-1 MPA--------------TDRFSDRATLARSVRLLTAFRFEQSAPDRFYGA
TH_2028|M.thermoresistible__bu MDAPR-----AAARRDVTELFARRATLRRSVRLLSEFRFEQSDPARFYGA
MMAR_0473|M.marinum_M --------------MAVTDRFARRATLSRSLRLLSEFRYEQPDPARFYGA
MUL_1123|M.ulcerans_Agy99 --------------MAVTDRFARRATLSRSLRLLSEFRYEQPDPARFYGA
MAV_4945|M.avium_104 ----------------MTDIFARRATLARSVRLLSQFRYERSEPARFYGA
Mb0229c|M.bovis_AF2122/97 --------------MAVTDVFARRATLRRSLRLLADFRYEQRDPARFYRT
Rv0224c|M.tuberculosis_H37Rv --------------MAVTDVFARRATLRRSLRLLADFRYEQRDPARFYRT
MLBr_02584|M.leprae_Br4923 MDTRRRVWPVAIITPHVTDVFAQRATLTRSLQLLSEFRYEQPYPARFYRA
MAB_4481|M.abscessus_ATCC_1997 -------------MPDITDLFAQRATLRRSTSLLTAFRFEQSAPERFYGT
MSMEG_6483|M.smegmatis_MC2_155 --------------------------------------------------
Mflv_0427|M.gilvum_PYR-GCK LASDTVAMVDDLWRATTGTSTRNLTVLDVGGGPGYFASAFTDAGLHYVGV
Mvan_0241|M.vanbaalenii_PYR-1 LADDTVAMVGDLWESATGESQAGLTLLDVGGGPGYFASAFTAAGVNYIGV
TH_2028|M.thermoresistible__bu LADDTAALVADLWRAATGSDPAGRTVLDVGGGPGYFAAAFARAGMRYIGV
MMAR_0473|M.marinum_M LAADTATLVSDLWLAARGESVAGRTLLDVGGGPGYFASAFAAAGVGYIGV
MUL_1123|M.ulcerans_Agy99 LAADTATLVSDLWLAARGESVAGRTLLDVGGGPGYFASAFAAAGVGYIGV
MAV_4945|M.avium_104 LAADTAAMVDDLWRAGHGESAAGRTLLDVGGGPGYFAAAFADAGVRYLGV
Mb0229c|M.bovis_AF2122/97 LAADTAAMIGDLWLATHSEPPVGRTLLDVGGGPGYFATAFSDAGVGYIGV
Rv0224c|M.tuberculosis_H37Rv LAADTAAMIGDLWLATHSEPPVGRTLLDVGGGPGYFATAFSDAGVGYIGV
MLBr_02584|M.leprae_Br4923 LAADTVAMVGDLWLSTHGAPPVGRILLDVGSGSGYFAAAFADAGVRYIGV
MAB_4481|M.abscessus_ATCC_1997 LAQDTVHLITDLWQQAEGVALRGRTVLDVGGGPGYFASAFADAGARYIGV
MSMEG_6483|M.smegmatis_MC2_155 ------MLTVDFDRLGVG---PGTSVIDVGCGAGRHTFEAFRRGAHVIGF
: *: . . ::*** *.* .: * :*.
Mflv_0427|M.gilvum_PYR-GCK EPDPGEMHAAAPRTHSR----------AGSFVRASGTALPFADGSVDICL
Mvan_0241|M.vanbaalenii_PYR-1 EPDPREMHAAAALTHER----------AGTFVRASGTALPFADDSVDVCL
TH_2028|M.thermoresistible__bu EPDPREIHAGPAVECPEDDP------AAGTFVRASGMALPFADASVDICL
MMAR_0473|M.marinum_M EPDPNEMHAAGPAHAGG----------PGSFVRASGMALPFADDSVDICL
MUL_1123|M.ulcerans_Agy99 EPDPNEMHAAGPAHAGG----------PGSFVRASGMALPLADDSVDICL
MAV_4945|M.avium_104 EPDPGEMHAAGPVVAAD----------TGTFVRASGMALPFADDSVDICL
Mb0229c|M.bovis_AF2122/97 EPDPDEMHAAGPAFTGR----------PGMFVRASGMALPLADDSVDICL
Rv0224c|M.tuberculosis_H37Rv EPDPDEMHAAGPAFTGR----------PGMFVRASGMALPFADDSVDICL
MLBr_02584|M.leprae_Br4923 EPDPREMHAAGPALVPK----------AGAFVRASGMALPFADDAVDICL
MAB_4481|M.abscessus_ATCC_1997 EPDPREMHAAPQGHRGIQDP-------RTTYLRASGMALPLADNSVDICL
MSMEG_6483|M.smegmatis_MC2_155 DQNVADLNSVDEMLQAMAEAGEAPATAKGEAVKGDALDLPYEDGTFDCVI
: : :::: ::... ** * :.* :
Mflv_0427|M.gilvum_PYR-GCK SSNVAEHVAQPWRLGEEMLRVTRPGGLAVLSYTVWLGPFGGHEMGLSHYL
Mvan_0241|M.vanbaalenii_PYR-1 SSNVAEHVAQPWRLGDEMLRVTRPGGLAVLSYTVWLGPFGGHEMGLTHYL
TH_2028|M.thermoresistible__bu SSNVAEHVPQPWLLGREMLRVTRPGGLAVLSYTVWLGPFGGHETGLWHYL
MMAR_0473|M.marinum_M SSNVAEHVPRPWQLGAEMLRVTRPGGLAVLSYTVWLGPFGGHEMGLSHYL
MUL_1123|M.ulcerans_Agy99 SSNVAEHVPRPWQLGAEMLRVTRPGGLAVLSYTVWLGPFGGHEMGLSHYL
MAV_4945|M.avium_104 SSNVAEHVPRPWQLGAEMLRVTRPGGLAVLSYTVWLGPFGGHEMGLTHYL
Mb0229c|M.bovis_AF2122/97 SSNVAEHVPRPWQLGTEMLRVTKPGGLVVLSYTVWLGPFGGHEMGLSHYL
Rv0224c|M.tuberculosis_H37Rv SSNVAEHVPRPWQLGTEMLRVTKPGGLVVLSYTVWLGPFGGHEMGLSHYL
MLBr_02584|M.leprae_Br4923 SSNVAEHVPQPWQLGNEMLRVTKPGGLVVLSYTIWLGPFGGHEMGLTHYL
MAB_4481|M.abscessus_ATCC_1997 SSNVAEHVPQPWRLGEEMLRVTRPGGLAILSYTVWLGPFGGHEMGLTHYF
MSMEG_6483|M.smegmatis_MC2_155 ASEILEHVPEDDRAISELVRVLKPGGSLAITVPRWLPEKICWLLSDEYHA
:*:: ***.. *::** :*** :: . ** . ::
Mflv_0427|M.gilvum_PYR-GCK GGYRAAERYTRRHGHRPKNDYGSSLFAVSAADGLR----WARSTG----A
Mvan_0241|M.vanbaalenii_PYR-1 GGRRAAEMYTRRHGHRPKNDYGSSLFAVHARDGLR----WAQGTG----A
TH_2028|M.thermoresistible__bu GGRRAADRYARRHGHRPKNDYGSSLFAVSAADGLR----WAAATG----A
MMAR_0473|M.marinum_M GGARAAARYARRHGHPAKNNYGSSLFAVSAAEGLR----WAEGTG----T
MUL_1123|M.ulcerans_Agy99 GGARAAARYARRHGHPAKNNYGSSLFAVSAAEGLR----WAEGTG----T
MAV_4945|M.avium_104 GGSRAAARYARKHGHPAKNNYGSSLFEVSVADGLA----WAACTG----A
Mb0229c|M.bovis_AF2122/97 GGARAAARYVRKHGHPAKNNYGSSLFAVSAAEGLR----WAAGTG----A
Rv0224c|M.tuberculosis_H37Rv GGARAAARYVRKHGHPAKNNYGSSLFAVSAAEGLR----WAAGTG----A
MLBr_02584|M.leprae_Br4923 GGARAATRYVRKHGHRAKNDYGSSLFPVSAADGLN----WAASTG----V
MAB_4481|M.abscessus_ATCC_1997 GGARAARWYTRKHGHPPKNNYGSSLFPVSVRDGLR----WARDTG----T
MSMEG_6483|M.smegmatis_MC2_155 NEGGHIRIYKADELRDKVLAHGLELTHTHHEHALHSPYWWLKCLVGTEKN
. * . : :* .* . ..* *
Mflv_0427|M.gilvum_PYR-GCK LAAAFPRYHPRWAWGLTRVPGVREFLVSNLVLVLRPT------------
Mvan_0241|M.vanbaalenii_PYR-1 LLAAFPRYHPRWAWGLTKVPGLREFLVSNLVLVLRPC------------
TH_2028|M.thermoresistible__bu LLAAFPRYHPKWGWWLTRAPVVREFLVSNLVLVLSPPEIVN--------
MMAR_0473|M.marinum_M LIAAFPRYHPRWAWWMTSVPMLREFLVSNLVLVLSPR------------
MUL_1123|M.ulcerans_Agy99 LIAAFPRYHPRSAWWMTSVPMLREFLVSNLVLVLSPR------------
MAV_4945|M.avium_104 ELAAFPRYHPRWAWWLTSVPVLREFLVSNLVLVLQPQ------------
Mb0229c|M.bovis_AF2122/97 ALAVFPRYHPRWAWWLTSVPVLREFLVSNLVLVLTP-------------
Rv0224c|M.tuberculosis_H37Rv ALAVFPRYHPRWAWWLTSVPVLREFLVSNLVLVLTP-------------
MLBr_02584|M.leprae_Br4923 AVAAFPRYHPRWAWWLTSVPVLREFVVSNLVLVLRPR------------
MAB_4481|M.abscessus_ATCC_1997 LTAAFPRYHPKWAWWITKIPALREVLVSNLVLVLRPR------------
MSMEG_6483|M.smegmatis_MC2_155 DHPAVKAYHQLLVWDMMGRPWLTRTAESALNPLIGKSVALYFTKPDARG
... ** * : * : . * * ::