For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
ADSKTARRVAILGGNRIPFARSDGAYAQASNQDMFTAVLDGLADRFNLKGEKLDAVIGGAVLKHSRDFNL MRECVLGSSLSSYTPAFDLQQACGTGLQSAIAAADGIARGRYDVAAAGGVDTASDAPIAFGDDLRKVLLG LRRAKSNVDRLKLVGKLPASLGVEIPVNSEPRTGMSMGEHAAVTAKEMGVKRVDQDELAAASHRNMSAAY DRGFFDDLVSPFLGLYRDNNLRPDSSAEKLAKLKPVFGVRNGDATMTAGNSTPLTDGASVALLSSEEWAA QHSIPVLAYLVDAETAAVDYVNGRDGLLMAPTYAVPRLLTRNGLTLQDFDYYEIHEAFASVVLATLAAWE SDEYCKERLGLDKALGSIDRSKLNVNGSSLAAGHPFAATGGRIVAQLAKQLAEKKKQTGQPVRGLISVCA AGGQGVTAILEA*
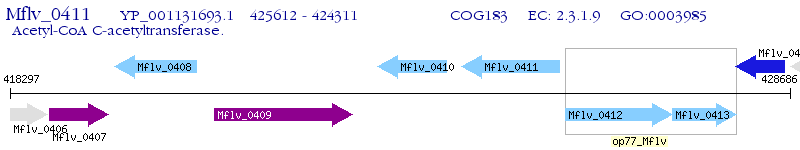
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0411 | - | - | 100% (433) | acetyl-CoA acetyltransferase |
| M. gilvum PYR-GCK | Mflv_4368 | - | 1e-43 | 31.41% (433) | acetyl-CoA acetyltransferase |
| M. gilvum PYR-GCK | Mflv_1347 | - | 9e-41 | 31.21% (439) | acetyl-CoA acetyltransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0249 | fadA2 | 0.0 | 84.72% (432) | acetyl-CoA acetyltransferase |
| M. tuberculosis H37Rv | Rv0243 | fadA2 | 0.0 | 84.72% (432) | acetyl-CoA acetyltransferase |
| M. leprae Br4923 | MLBr_02564 | fadA2 | 0.0 | 79.44% (428) | acetyl-CoA acetyltransferase |
| M. abscessus ATCC 19977 | MAB_4442c | - | 0.0 | 81.94% (432) | acetyl-CoA acetyltransferase |
| M. marinum M | MMAR_0504 | fadA2 | 0.0 | 82.72% (434) | acetyl-CoA acyltransferase FadA2 |
| M. avium 104 | MAV_4915 | - | 0.0 | 83.61% (427) | acetyl-CoA acetyltransferase |
| M. smegmatis MC2 155 | MSMEG_0373 | - | 0.0 | 87.44% (430) | acetyl-CoA acetyltransferase |
| M. thermoresistible (build 8) | TH_0166 | fadA6 | 6e-34 | 28.11% (434) | PROBABLE ACETYL-CoA ACETYLTRANSFERASE FADA6 |
| M. ulcerans Agy99 | MUL_1167 | fadA2 | 0.0 | 82.72% (434) | acetyl-CoA acetyltransferase |
| M. vanbaalenii PYR-1 | Mvan_0260 | - | 0.0 | 93.53% (433) | acetyl-CoA acetyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0411|M.gilvum_PYR-GCK ----------------------MADSKTARRVAILGGNRIPFARSDGAYA
Mvan_0260|M.vanbaalenii_PYR-1 ----MTLDTPSVSR-----EADVADTNTTRRVAILGGNRIPFARADGAYA
MSMEG_0373|M.smegmatis_MC2_155 ----------------------MAN-ESRRRVAVLGGNRIPFARSDGAYA
MAB_4442c|M.abscessus_ATCC_199 ---------MATSS-----ARKSDASQGRRRVAVLGGNRIPFARSDGAYA
Mb0249|M.bovis_AF2122/97 ---------MAPAA--------KNTSQTRRRVAVLGGNRIPFARSDGAYA
Rv0243|M.tuberculosis_H37Rv ---------MAPAA--------KNTSQTRRRVAVLGGNRIPFARSDGAYA
MMAR_0504|M.marinum_M ---------MAPAA--------EKNAQTKRRVAVLGGNRIPFARSDGAYA
MUL_1167|M.ulcerans_Agy99 ---------MAPAA--------EKNAQTKRRVAVLGGNRIPFARSDGAYA
MAV_4915|M.avium_104 -------------------------------MAVLGGNRIPFARSDGAYA
MLBr_02564|M.leprae_Br4923 MLGETRRLQVAPASSVASDKTIQRTTFAKQRVAVLGGNRIPFARSNGAYA
TH_0166|M.thermoresistible__bu --------------------------MAMTEAYIVDAVRTPVGKRNGGLS
::.. * *..: :*. :
Mflv_0411|M.gilvum_PYR-GCK QASNQDMFTAVLDGLADRFNLKGEKLDAVIGGAVLKH-SRDFNLMRECVL
Mvan_0260|M.vanbaalenii_PYR-1 KASNQDMFTATLDGLADRFNLKGEKLDAVIGGAVLKH-SRDFNLMRECVL
MSMEG_0373|M.smegmatis_MC2_155 NASNQDMFTAALGGLIDRFNLAGEQLGAVIGGAVLKH-SRDFNLMRECVL
MAB_4442c|M.abscessus_ATCC_199 NASNQDMLTAAITGASDRFNLKGERLGAVIAGAVLKL-SRDFNLTRESVL
Mb0249|M.bovis_AF2122/97 DASNQDMFTAALSGLVDRFGLAGERLDMVVGGAVLKH-SRDFNLMRECVL
Rv0243|M.tuberculosis_H37Rv DASNQDMFTAALSGLVDRFGLAGERLDMVVGGAVLKH-SRDFNLMRECVL
MMAR_0504|M.marinum_M EASNQDMFTAALAGLVDRFDLRGQRLGAVVGGAVLKH-SRDFNLMRECVL
MUL_1167|M.ulcerans_Agy99 EASNQDMFTAALAGLVDRFDLRGQRLGAVVGGAVLKH-SRDFNLMRECVL
MAV_4915|M.avium_104 EASNQDMLTAALEGLVDRFGLAGERLGVVVGGAVLKH-SRDFNLTRECVL
MLBr_02564|M.leprae_Br4923 EASNQDMFSAALSGLVDRFGLAGERLDMVIGGAVLKH-SRDFNLMRECVL
TH_0166|M.thermoresistible__bu GAHPADMAAHVIKTLVGRHDFDPAAIDDVIFGCLDNIGSQAGDIARTAAL
* ** : .: .*..: :. *: *.: : *: :: * ..*
Mflv_0411|M.gilvum_PYR-GCK GSSLSSYTPAFDLQQACGTGLQSAIAAADGIARGRYDVAAAGGVDTASDA
Mvan_0260|M.vanbaalenii_PYR-1 GSSLSSATPAFDLQQACGTGLQAAIAAADGIARGRYEVAAAGGVDTASDA
MSMEG_0373|M.smegmatis_MC2_155 GSALSPYTPAFDIQQACGTGLQAAIAAADGIAAGRYESAAAGGVDTASDA
MAB_4442c|M.abscessus_ATCC_199 GSPLSPYTQAFDLQQACGTGLQAAIVGADGIALGRYESVLAGGVDTTSDA
Mb0249|M.bovis_AF2122/97 GSELSPYTPAFDLQQACGTGLQAAIAAADGIAAGRYEVAAAGGVDTTSDP
Rv0243|M.tuberculosis_H37Rv GSELSPYTPAFDLQQACGTGLQAAIAAADGIAAGRYEVAAAGGVDTTSDP
MMAR_0504|M.marinum_M GSELSPYTAACDLQQACGTGLQAAIAAADGIAAGRYEVAAAGGVDTTSDP
MUL_1167|M.ulcerans_Agy99 GSELSPYTAACDLQQACGTGLQAAIAAADGIAAGRYEVAAAGGVDTTSDP
MAV_4915|M.avium_104 GSQLSSYTPAFDLQQACGTGLQAAIAAAEGIAAGRYDVAAAGGVDTTSDA
MLBr_02564|M.leprae_Br4923 GSQLSPYTPAFDLQQACATGLQAAIVAADGIAAGRYEVAVAGGVDTTSDV
TH_0166|M.thermoresistible__bu AAGLPESVPGVTIDRQCGSAQQAVHFAAQGVMSGTVDLIVAGGVQKMSQF
.: *. . . ::: *.:. *:. .*:*: * : ****:. *:
Mflv_0411|M.gilvum_PYR-GCK PIAFGDDLRKVLLGLRRAKSNVDRLKLVGKLPASLGVEIPVNSEPRTGMS
Mvan_0260|M.vanbaalenii_PYR-1 PIAFGDDLRRVLLGLRRAKSNVERLKLVGKLPAALGVEIPVNSEPRTGMS
MSMEG_0373|M.smegmatis_MC2_155 PIAFGNDLRRVLLGLRRAKSNVDRLKLVGKLPASLGVEIPVNSEPRTGMS
MAB_4442c|M.abscessus_ATCC_199 PINFGDDLRHTLLAIRRAKDNVTRLKLLGKLPAALGVDIPQNGEPRTGLS
Mb0249|M.bovis_AF2122/97 PIGLGDDLRRTLLKLRRSRSNVQRLKLVGTLPASLGVEIPANSEPRTGLS
Rv0243|M.tuberculosis_H37Rv PIGLGDDLRRTLLKLRRSRSNVQRLKLVGTLPASLGVEIPANSEPRTGLS
MMAR_0504|M.marinum_M PIGLGEDLRRTLLRLRRAKSNVHRLRLVGGLPANLGVEIPANSEPRTGLS
MUL_1167|M.ulcerans_Agy99 PIGLGEDLRRTLLRLRRAKSNVHRLRLVGGLPANLGVEIPANSEPRTGLS
MAV_4915|M.avium_104 PIGLGDNLRRTLLKLRRAKSNVQRLKLVGTLPATLGVEIPVNSEPRTGLS
MLBr_02564|M.leprae_Br4923 PISLGDNLRRNLLKLRRSKSNVQRLKLVGALPANIGVEIPANSEPRTGLS
TH_0166|M.thermoresistible__bu PILS----------------AFAAGEPYGATDPWTGCEGWQARYGDQEIS
** . . * . * : :*
Mflv_0411|M.gilvum_PYR-GCK MGEHAAVTAKEMGVKRVDQDELAAASHRNMSAAYDRGFFDDLVSPFLGLY
Mvan_0260|M.vanbaalenii_PYR-1 MGEHAAVTAKEMGVKRVDQDELAAASHRNMAAAYDRGFFDDLVTPFLGLY
MSMEG_0373|M.smegmatis_MC2_155 MGEHAAVTAKAMGIKRVEQDELAAASHRNMAAAYDAGFFDDLVTPFLGVY
MAB_4442c|M.abscessus_ATCC_199 MGEHAAITAKQAGVKRTEQDALAAASHQKMAAAYDRGFFDDLISPFLGLY
Mb0249|M.bovis_AF2122/97 MGEHAAVTAKQMGIKRVDQDELAAASHRNMADAYDRGFFDDLVSPFLGLY
Rv0243|M.tuberculosis_H37Rv MGEHAAVTAKQMGIKRVDQDELAAASHRNMADAYDRGFFDDLVSPFLGLY
MMAR_0504|M.marinum_M MGDHAAITAKQMGIKRVDQDELAAASHRNMADAYDRGFFDDLVTPFLGLY
MUL_1167|M.ulcerans_Agy99 MGDHAAITAKQMGIKRVDQDELAAASHRNMADAYDRGFFDDLVTPFLGLY
MAV_4915|M.avium_104 MGEHQAITAKQMGITRVAQDELAAASHRNMAAAYDRGFFDDLVTPFLGLY
MLBr_02564|M.leprae_Br4923 MGEHAAITAKKMGIKRVDQDELAAASHRNMASAYDRGFFDDLVTPFLGLY
TH_0166|M.thermoresistible__bu QFRGAEMIAEHWKLSREDNEKFALESHRRAVAAIEAGYFDKEITPYADAT
: *: :.* :: :* **:. * : *:**. ::*: .
Mflv_0411|M.gilvum_PYR-GCK RDNNLRPDSSAEKLAKLKPVFGVRNGDATMTAGNSTPLTDGASVALLSSE
Mvan_0260|M.vanbaalenii_PYR-1 RDNNLRADSSVEKLAKLRPVFGVRSGDATMTAGNSTPLTDGASVALLSSE
MSMEG_0373|M.smegmatis_MC2_155 RDNNLRADSSVEKLAKLKPVFGVKAGDATMTAGNSTPLTDGASVALLAGE
MAB_4442c|M.abscessus_ATCC_199 RDDNLRPNSSVEKLATLKPVFGVKHGDATMTAGNSTPLTDGASVALLSSE
Mb0249|M.bovis_AF2122/97 RDDNLRPNSSVEKLATLRPVFGVKAGDATMTAGNSTPLTDGASVALLASE
Rv0243|M.tuberculosis_H37Rv RDDNLRPNSSVEKLATLRPVFGVKAGDATMTAGNSTPLTDGASVALLASE
MMAR_0504|M.marinum_M RDDNLRPNSSVEKLASLRPVFGVKAGDATMTAGNSTPLTDGASVALLSSE
MUL_1167|M.ulcerans_Agy99 RDDNLRPNSSVEKLASLRPVFGVKAGDATMTAGNSTPLTDGASVALLSSE
MAV_4915|M.avium_104 RDDNLRPDSSPEKLAKLRPVFGVKAGDATMTAGNSTPLTDGASVALLGTE
MLBr_02564|M.leprae_Br4923 RDDNLRPNSHVEKLATLRPVFGVQAGDATMTAGNSTSLTDGASVALLATE
TH_0166|M.thermoresistible__bu VDEGPRPDTSLEKMASLPTLIEG----GVLTAAVASQISDGAAALLVASK
*:. *.:: **:*.* .:: ..:**. :: ::***:. *:. :
Mflv_0411|M.gilvum_PYR-GCK EWAAQHSIPVLAYLVDAETAAVDYVNGRDGLLMAPTYAVPRLLTRNGLTL
Mvan_0260|M.vanbaalenii_PYR-1 EWAAEHSIPVLAYFVDGQTAAVDYVNGRDGLLMAPTYAVPRLLARNGLTL
MSMEG_0373|M.smegmatis_MC2_155 DWAQARGLEPLAYFVDAETAAVDYVNGPDGLLMAPTYAVPRLLARNNLTL
MAB_4442c|M.abscessus_ATCC_199 EWAAERNIPVLAYFVDGETAAVDYVNGPDGLLMAPTYAVPRLLARNGLTL
Mb0249|M.bovis_AF2122/97 QWAEAHSLAPLAYLVDAETAAVDYVNGNDGLLMAPTYAVPRLLARNGLSL
Rv0243|M.tuberculosis_H37Rv QWAEAHSLAPLAYLVDAETAAVDYVNGNDGLLMAPTYAVPRLLARNGLSL
MMAR_0504|M.marinum_M QWAQEHSLSPLAYLVDAETAAVDYVNGNDGLLMAPTYAVPRLLARNGLTL
MUL_1167|M.ulcerans_Agy99 QWAQEHSLSPLAYLVDAETAAVDYVNGNDGLLMAPTYAVPRLLARNGLTL
MAV_4915|M.avium_104 EWAAAHSLTPLAYLVDAETAAVDYVNGRDGLLMAPTYAVPRLLARNGLSL
MLBr_02564|M.leprae_Br4923 QWVAAHSLTPFAYWVDAEIAAVDYVSGNDGLLMAPTYAVPRLLARNGLSF
TH_0166|M.thermoresistible__bu DAVDRFNLTPRARIHHMSVRGADPVM----MLTAPIPATQHALKRAGMSI
: . .: * . . ..* * :* ** *. : * * .:::
Mflv_0411|M.gilvum_PYR-GCK QDFDYYEIHEAFASVVLATLAAWESDEYCKERLGLDKALGSIDRSKLNVN
Mvan_0260|M.vanbaalenii_PYR-1 QDFDFYEIHEAFASVVLATLAAWKSDEYCKERLGLDKALGSIDRSKLNVN
MSMEG_0373|M.smegmatis_MC2_155 QDFDFYEIHEAFASVVLAHLQAWESDEYCKERLGLDKALGSIDRTKLNVN
MAB_4442c|M.abscessus_ATCC_199 QDFDFYEIHEAFASVVLATLQAWESADYCKERLGLDKPLGAIDRSKLNVN
Mb0249|M.bovis_AF2122/97 QDFDFYEIHEAFASVVLAHLAAWESEEYCKRRLGLDAALGSIDRSKLNVN
Rv0243|M.tuberculosis_H37Rv QDFDFYEIHEAFASVVLAHLAAWESEEYCKRRLGLDAALGSIDRSKLNVN
MMAR_0504|M.marinum_M QDFDFYEIHEAFASVVLAHLQAWESEEYCKQRLGLDAALGSIDRSKLNVN
MUL_1167|M.ulcerans_Agy99 QDFDFYEIHEAFASVVLAHLQAWESEEYCKQRLGLDAALGSIDRSKLNVN
MAV_4915|M.avium_104 QDFDFYEIHEAFASVVLAHLQAWESEDYCKERLGLDAALGSIDRSKLNVN
MLBr_02564|M.leprae_Br4923 QDFDFYEIHEAFASVVLAHLQAWESEEYCRQCLGLDTALGPIDRSKLNVN
TH_0166|M.thermoresistible__bu DDIDAVEINEAFAPVVLAWLAELG-----------------ADPARVNPN
:*:* **:****.**** * * :::* *
Mflv_0411|M.gilvum_PYR-GCK GSSLAAGHPFAATGGRIVAQLAKQLAEKKK-QTG--QPVRGLISVCAAGG
Mvan_0260|M.vanbaalenii_PYR-1 GSSLAAGHPFAATGGRIVAQLAKQLAEKKA-ETG--KPVRGLISVCAAGG
MSMEG_0373|M.smegmatis_MC2_155 GSSLAAGHPFAATGGRIVAQLAKQLAEKKK-STG--RPVRGLISICAAGG
MAB_4442c|M.abscessus_ATCC_199 GSSLAAGHPFAATGGRIVAQLAKQLAEKKK-ENGGTGTYRGLISVCAAGG
Mb0249|M.bovis_AF2122/97 GSSLAAGHPFAATGGRILAQTAKQLAEKKAAKKG-GGPLRGLISICAAGG
Rv0243|M.tuberculosis_H37Rv GSSLAAGHPFAATGGRILAQTAKQLAEKKAAKKG-GGPLRGLISICAAGG
MMAR_0504|M.marinum_M GSSLAAGHPFAATGGRILAQTAKQLAEKKAERKGSAQPVRALISICAAGG
MUL_1167|M.ulcerans_Agy99 GSSLAAGHPFAATGGRILAQTAKQLAEKKAERKGSAQPVRALISICAAGG
MAV_4915|M.avium_104 GSSLAAGHPFAATGGRILAQAAKQVAQRKAELKGADKPVRALISICAAGG
MLBr_02564|M.leprae_Br4923 GSSLAAGHPFAATGGRILVQTAKQLAEQKATQKG-CGPMRALISVCAAGG
TH_0166|M.thermoresistible__bu GGAIALGHPIGCTGARLMTSLLHHLERTGG--------RYGLQTMCEGGG
*.::* ***:..**.*::.. ::: . .* ::* .**
Mflv_0411|M.gilvum_PYR-GCK QGVTAILEA-
Mvan_0260|M.vanbaalenii_PYR-1 QGVTAILEA-
MSMEG_0373|M.smegmatis_MC2_155 QGVAAILEA-
MAB_4442c|M.abscessus_ATCC_199 QGVTAILEA-
Mb0249|M.bovis_AF2122/97 QGVAAILEA-
Rv0243|M.tuberculosis_H37Rv QGVAAILEA-
MMAR_0504|M.marinum_M QGVAAILEA-
MUL_1167|M.ulcerans_Agy99 QGVAAILEA-
MAV_4915|M.avium_104 QGVAAILEA-
MLBr_02564|M.leprae_Br4923 QGVAVILEA-
TH_0166|M.thermoresistible__bu QANVTIIERL
*. ..*:*