For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VSSVDFRVFVEPQQGASYADQLAVARTAEQAGYSAFFRSDHYLAMAGDGLPGPTDSWVTLAGLARETSSI RLGTMVTSATFRHPGPLAISVAQVDEMSGGRVELGLGAGWFEAEHKAYAIPFPPLGERFDRLTEQLEILT GLWTTPTGETFDYSGTHYSIADSPALPKPAQSPRPPIVIGGRGAKRTPALTARFADEFNIPFVPLDELTT QYARVAAAVEDAGRAKDSLTYSAAFVVCVGRDDAEIARRAAAIGREVDEMRSNSPTVGTPAEVVDKLGPF LEAGVERVYLQVLDMSDLEHVEFFAEQVIPQIH
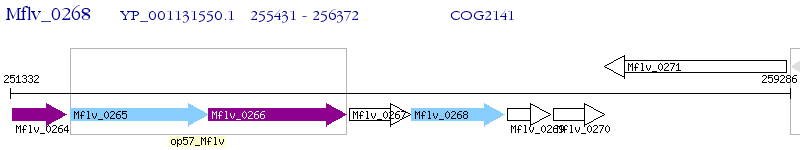
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0268 | - | - | 100% (313) | luciferase family protein |
| M. gilvum PYR-GCK | Mflv_4098 | - | 1e-44 | 34.41% (311) | luciferase family protein |
| M. gilvum PYR-GCK | Mflv_1117 | - | 1e-36 | 35.74% (249) | luciferase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0815c | - | 3e-41 | 30.15% (325) | hypothetical protein Mb0815c |
| M. tuberculosis H37Rv | Rv0791c | - | 3e-41 | 30.15% (325) | hypothetical protein Rv0791c |
| M. leprae Br4923 | MLBr_00348 | - | 8e-11 | 27.07% (229) | putative coenzyme F420-dependent oxidoreductase |
| M. abscessus ATCC 19977 | MAB_4277 | - | 1e-133 | 72.49% (309) | luciferase-like monooxygenase superfamily protein |
| M. marinum M | MMAR_5411 | - | 8e-40 | 38.55% (249) | hypothetical protein MMAR_5411 |
| M. avium 104 | MAV_0169 | - | 2e-37 | 37.35% (249) | hypothetical protein MAV_0169 |
| M. smegmatis MC2 155 | MSMEG_0702 | - | 1e-149 | 81.17% (308) | monooxygenase |
| M. thermoresistible (build 8) | TH_2824 | - | 1e-148 | 79.61% (309) | PUTATIVE luciferase family protein |
| M. ulcerans Agy99 | MUL_5034 | - | 2e-38 | 38.15% (249) | hypothetical protein MUL_5034 |
| M. vanbaalenii PYR-1 | Mvan_0629 | - | 1e-159 | 88.31% (308) | luciferase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0815c|M.bovis_AF2122/97 MNAKDDPHFGLMLAATVNGLAVGSYREMVVVSQTAEEYG-FDSVWLCDHF
Rv0791c|M.tuberculosis_H37Rv MNAKDDPHFGLMLAATVNGLAVGSYREMVVVSQTAEEYG-FDSVWLCDHF
MMAR_5411|M.marinum_M -----------MRFAFKTSPQNTTWEQMLAVWQTADGIDVYESGWTFDHF
MUL_5034|M.ulcerans_Agy99 -----------MRFAFKNSPQNTTWEQMLAVWQTADGIDVYESGWTFDHF
MAV_0169|M.avium_104 -----------MRFAFKTSPQNTTWAQMLAVWQAADDIDVYESGWTFDHF
Mflv_0268|M.gilvum_PYR-GCK -MSSV------DFRVFVEPQQGASYADQLAVARTAEQAG-YSAFFRSDHY
Mvan_0629|M.vanbaalenii_PYR-1 -MSTVAATNTPDFRIFVEPQQGASYADQLAVARTAESAG-YSAFFRSDHY
MSMEG_0702|M.smegmatis_MC2_155 ----------MDFRVFVEPQQGATYSDQLAVAQAAERLG-YSAFFRSDHY
TH_2824|M.thermoresistible__bu ----------MDFRVFVEPQQGGTYADQLAVARAAEDLG-YSAFFRSDHY
MAB_4277|M.abscessus_ATCC_1997 ----------MDFRVFVEPQQGATYSDQLRVAQAAEELG-FSAFFRSDHY
MLBr_00348|M.leprae_Br4923 ---MVERANAMKLGLQLGYWAAQPPGNAAELVAAAEGAG-FDAVFTGEAW
. : : :*: . :.: : : :
Mb0815c|M.bovis_AF2122/97 LTISPGEYAKVAGIAADTGSATGTETGGAGQCAPSRSLPLLECWTALAAL
Rv0791c|M.tuberculosis_H37Rv LTISPGEYAKVAGIAADTGSATGTETGGAGQCAPSRSLPLLECWTALAAL
MMAR_5411|M.marinum_M YPIFS-----------------------------DPTGPCLEGWTTLTAL
MUL_5034|M.ulcerans_Agy99 YPIFS-----------------------------DPTGPCLEGWTTLTAL
MAV_0169|M.avium_104 YPIFS-----------------------------DSSGPCLEGWTTLTAL
Mflv_0268|M.gilvum_PYR-GCK LAMAG-----------------------------DGLPGPTDSWVTLAGL
Mvan_0629|M.vanbaalenii_PYR-1 VAMAG-----------------------------DGLPGPTDSWVTLAGI
MSMEG_0702|M.smegmatis_MC2_155 LKMAG-----------------------------DGLPGPTDSWVTLAGI
TH_2824|M.thermoresistible__bu LAMSG-----------------------------DGLPGPTDSWVTLAGI
MAB_4277|M.abscessus_ATCC_1997 LAMGGA----------------------------DGLPGPTDAWVTLGAI
MLBr_00348|M.leprae_Br4923 G---------------------------------------SDAYTPLAWW
: :..*
Mb0815c|M.bovis_AF2122/97 SRDTTKLRLGTSVLCNSYRHPSVLAKMAATLDVISQGRLDLGLGAGWFRR
Rv0791c|M.tuberculosis_H37Rv SRDTTKLRLGTSVLCNSYRHPSVLAKMAATLDVISQGRLDLGLGAGWFRR
MMAR_5411|M.marinum_M AQATTRLRLGTLVTGVHYRHPAVLANMAAALDIISHGRLELGIGAGWNEE
MUL_5034|M.ulcerans_Agy99 AQATTRLRLGTLVTGIHYRHPAVLANMAAALDIISHGRLELGIGAGWNEE
MAV_0169|M.avium_104 AQATKRLRLGTLVTGIHYRHPAVLANMAAAVDIISGGRLELGIGAGWNEE
Mflv_0268|M.gilvum_PYR-GCK ARETSSIRLGTMVTSATFRHPGPLAISVAQVDEMSGGRVELGLGAGWFEA
Mvan_0629|M.vanbaalenii_PYR-1 ARETSTIRLGTMVTSATFRYPGPLAISVAQVDEMSGGRVEFGIGAGWFEA
MSMEG_0702|M.smegmatis_MC2_155 ARETSTIRLGTMVTSATFRYPGPLAISVAQVDEMSGGRVELGLGAGWFEE
TH_2824|M.thermoresistible__bu ARETTSIRLGTLVTSATFRYPGPLAISVAQVDEMSGGRVEFGLGSGWFEP
MAB_4277|M.abscessus_ATCC_1997 ARETTAIRLGTLVTSATFRHPGPLAVTVAQVDEMSSGRVDFGLGAGWFSE
MLBr_00348|M.leprae_Br4923 GSSTQRVRLGTSVVQLSARTPTACAMAALTLDHLSGGRHILGLGVSGPQV
. * :**** * * * * . :* :* ** :*:* .
Mb0815c|M.bovis_AF2122/97 ESQAYGIPFPPVGDRVSALAESLQVIKAVWTEP---NPTYAGRFYTLDGA
Rv0791c|M.tuberculosis_H37Rv ESQAYGIPFPPVGDRVSALAESLQVIKAVWTEP---NPTYAGRFYTLDGA
MMAR_5411|M.marinum_M ESSAYGIELGSVKERFDRFEEACAVLTGLLSQP---STTFDGKYYQLEDA
MUL_5034|M.ulcerans_Agy99 ESGAYGIELGSVKERFDRFEEACAVLTGLLSQP---STTFDGKYYQLEDA
MAV_0169|M.avium_104 ESGAYGIELGSIRERFDRFEEACAVLTSLLSQE---TTDFDGKFYQLKGA
Mflv_0268|M.gilvum_PYR-GCK EHKAYAIPFPPLGERFDRLTEQLEILTGLWTTPTGETFDYSGTHYSIADS
Mvan_0629|M.vanbaalenii_PYR-1 EHKAYAIPFPPLGERFDRLTEQLEILTGLWTAAPGETFDYSGRHYSIVDS
MSMEG_0702|M.smegmatis_MC2_155 EHLAYAIPFPPLGERFDRLTEQLQILTGMWGTPVGDTFDFAGTHYTVKDS
TH_2824|M.thermoresistible__bu EHQAYAIPFPPLGERFDRLTEQLEIITGLWTTPSGEKYDFTGKHYTVVDS
MAB_4277|M.abscessus_ATCC_1997 EHEAYAIPFPSLGERFDRLEEQLEILTGLWTTPTGDTYSYAGKHYTVTDS
MLBr_00348|M.leprae_Br4923 VEGWYGQPFP---KPLSRTREYIDIVRQVWAREAPVVSDGQHYPLPLTGA
*. : . .. * :: : : .:
Mb0815c|M.bovis_AF2122/97 TCD-----PPPVQRPH---PPLWIGGEGD-RVQRIAAKHAQGLNVRWWSP
Rv0791c|M.tuberculosis_H37Rv TCD-----PPPVQRPH---PPLWIGGEGD-RVQRIAAKHAQGLNVRWWSP
MMAR_5411|M.marinum_M RNE-----PKGPQRPH---PPICIGGSGEKRTLRITARYAQHWNFAGGTP
MUL_5034|M.ulcerans_Agy99 RNE-----PKGPQRPH---SPICIGGSGEKRTLRITARYAQHWNFAGGTP
MAV_0169|M.avium_104 RNE-----PKGPQRPH---PPICIGGSGEKRTLRITARYAQHWNFVGGPP
Mflv_0268|M.gilvum_PYR-GCK PAL-----PKPAQSPR---PPIVIGGRGAKRTPALTARFADEFNIPFVPL
Mvan_0629|M.vanbaalenii_PYR-1 PAL-----PKPAQRPH---PPIIIGGGGAKRTPALAARFADEFNIPFVPL
MSMEG_0702|M.smegmatis_MC2_155 PGL-----PKPVQTPH---PPIIIGGQGAKRTPALAAQFASEFNIPFVPL
TH_2824|M.thermoresistible__bu PGL-----PKPVQDPY---PPIIIGGQGPKRTPALAARFANEFNIPFVRL
MAB_4277|M.abscessus_ATCC_1997 PAL-----PKPVQEPH---PPIVIGGLGVKRTPALAARFATEFNLPFQPV
MLBr_00348|M.leprae_Br4923 GATGLGKALKPITHPLRADIPIMLGAEGP-KNVALAAEICDGWLPIFFSP
* *: :*. * : ::*. .
Mb0815c|M.bovis_AF2122/97 QQVTQRRGFLTQASEAAGRDPD----TLRLSVTLLLAPTQS---------
Rv0791c|M.tuberculosis_H37Rv QQVTQRRGFLTQASEAAGRDPD----TLRLSVTLLLAPTQS---------
MMAR_5411|M.marinum_M EEFARKRDVLAAHCADIGRDPK----EITLSAHLGLAPER----------
MUL_5034|M.ulcerans_Agy99 EEFARKRDVLAAHCADIGRDPK----EITLSAHRGLAPER----------
MAV_0169|M.avium_104 DVFAHKRDVLASHCADIGRDPK----EILLSAHLRLEPDL----------
Mflv_0268|M.gilvum_PYR-GCK DELTTQYARVAAAVEDAGRAKD----SLTYSAAFVVCVGRDDAEI-----
Mvan_0629|M.vanbaalenii_PYR-1 DTLTEQYGRVAAAVEQAGRAKD----SLIYSAAFVLCVGRDETEI-----
MSMEG_0702|M.smegmatis_MC2_155 DTLKTQFERVAAAVEGAGRSAD----SMIYSAAFVLCAGADEAQI-----
TH_2824|M.thermoresistible__bu DALKAQFERVAAAVAEAGREPD----SMIYSAAFVLCAGRDDAEI-----
MAB_4277|M.abscessus_ATCC_1997 DVITDQYARVAHAVEAAGRPAD----SMTYSAAFALCIGDTDADI-----
MLBr_00348|M.leprae_Br4923 RMADMYNEWLDEGFARTGARRSREDFEICASAQIVVTEDRAAAFAGIKPF
: * . : *. :
Mb0815c|M.bovis_AF2122/97 -GEEEVRIREEFASIPEPGLIVGTPDRCVERIREYQDRGVGHFLFTIPHV
Rv0791c|M.tuberculosis_H37Rv -GEEEVRIREEFASIPEPGLIVGTPDRCVERIREYQDRGVGHFLFTIPHV
MMAR_5411|M.marinum_M ---DFGQVIENAAALAAEGLDLG-----IVYIAPPHD----------PAV
MUL_5034|M.ulcerans_Agy99 ---DFGQVIENAAALAAEGLDLG-----IVYIAPPHD----------PAV
MAV_0169|M.avium_104 ---DYGKVIDDAAALAAEGLDLG-----IVYLPPPHD----------PAV
Mflv_0268|M.gilvum_PYR-GCK -ARRAAAIGREVDEMRSNSPTVGTPAEVVDKLGPFLEAGVERVYLQVLDM
Mvan_0629|M.vanbaalenii_PYR-1 -ARRAAAIGREVDELRSNSPVVGTPGEVVDKLRPFLEAGVQRIYLQLLDM
MSMEG_0702|M.smegmatis_MC2_155 -AKRAAAIGREVDELRSNSPLVGTSTEIVDKLGPFIEAGVQRVYLQVLDM
TH_2824|M.thermoresistible__bu -ARRAAAINREVDELRSNSPVVGTPAEIVDRLGPFIEAGVQRVYLQLLDM
MAB_4277|M.abscessus_ATCC_1997 -ARRAANIGREVEEITENSPLAGTPDAVAEKLSIYVDAGVQRVYVQLLDI
MLBr_00348|M.leprae_Br4923 LALYMGGMGAEGTNFHADVYRRMGYAEVVDEVTALFRSNQKDKAAEIIPN
: : : :
Mb0815c|M.bovis_AF2122/97 VKSDYLHII-----------------------GSDIIPRVKTEVTIP
Rv0791c|M.tuberculosis_H37Rv VKSDYLHII-----------------------GSDIIPRVKTEVTIP
MMAR_5411|M.marinum_M LEPMAEAIR-----------------------ESGLIDG--------
MUL_5034|M.ulcerans_Agy99 LEPMAEAIR-----------------------ESGLIDG--------
MAV_0169|M.avium_104 LEPLAEAIR-----------------------DSGLLRS--------
Mflv_0268|M.gilvum_PYR-GCK SDLEHVEFF-----------------------AEQVIPQIH------
Mvan_0629|M.vanbaalenii_PYR-1 SDLDHVQFF-----------------------AEQVIPRIG------
MSMEG_0702|M.smegmatis_MC2_155 SDLDHVEFF-----------------------ANEVVRQFS------
TH_2824|M.thermoresistible__bu SDLDHLELI-----------------------ATQVVPQLR------
MAB_4277|M.abscessus_ATCC_1997 RDLDHLEFF-----------------------ASTVIPQFS------
MLBr_00348|M.leprae_Br4923 ELVDSTMIVGDVDYVCKQIAAWSAAGVTMMMVSASSVEQVRDLAGLF
: :