For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
DFRVFVEPQQGATYSDQLAVAQAAERLGYSAFFRSDHYLKMAGDGLPGPTDSWVTLAGIARETSTIRLGT MVTSATFRYPGPLAISVAQVDEMSGGRVELGLGAGWFEEEHLAYAIPFPPLGERFDRLTEQLQILTGMWG TPVGDTFDFAGTHYTVKDSPGLPKPVQTPHPPIIIGGQGAKRTPALAAQFASEFNIPFVPLDTLKTQFER VAAAVEGAGRSADSMIYSAAFVLCAGADEAQIAKRAAAIGREVDELRSNSPLVGTSTEIVDKLGPFIEAG VQRVYLQVLDMSDLDHVEFFANEVVRQFS*
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
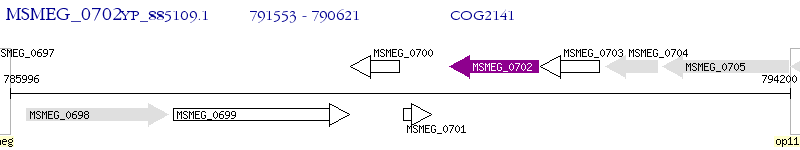
| M. smegmatis MC2 155 | MSMEG_0702 | - | - | 100% (310) | monooxygenase |
| M. smegmatis MC2 155 | MSMEG_3301 | - | 5e-37 | 35.63% (247) | hypothetical protein MSMEG_3301 |
| M. smegmatis MC2 155 | MSMEG_6454 | - | 1e-36 | 34.94% (249) | hypothetical protein MSMEG_6454 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0815c | - | 1e-38 | 29.85% (325) | hypothetical protein Mb0815c |
| M. gilvum PYR-GCK | Mflv_0268 | - | 1e-149 | 81.17% (308) | luciferase family protein |
| M. tuberculosis H37Rv | Rv0791c | - | 1e-38 | 29.85% (325) | hypothetical protein Rv0791c |
| M. leprae Br4923 | MLBr_00348 | - | 9e-12 | 29.41% (238) | putative coenzyme F420-dependent oxidoreductase |
| M. abscessus ATCC 19977 | MAB_4277 | - | 1e-133 | 72.67% (311) | luciferase-like monooxygenase superfamily protein |
| M. marinum M | MMAR_2559 | - | 5e-39 | 31.65% (316) | oxidoreductase |
| M. avium 104 | MAV_0169 | - | 5e-39 | 37.35% (249) | hypothetical protein MAV_0169 |
| M. thermoresistible (build 8) | TH_2824 | - | 1e-148 | 82.79% (308) | PUTATIVE luciferase family protein |
| M. ulcerans Agy99 | MUL_3202 | - | 2e-38 | 31.33% (316) | oxidoreductase |
| M. vanbaalenii PYR-1 | Mvan_0629 | - | 1e-146 | 81.31% (305) | luciferase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0815c|M.bovis_AF2122/97 MNAKDD-------PHFGLMLAATVNGLAVGSYREMVVVSQTAEEYG-FDS
Rv0791c|M.tuberculosis_H37Rv MNAKDD-------PHFGLMLAATVNGLAVGSYREMVVVSQTAEEYG-FDS
MMAR_2559|M.marinum_M MHENTSNPPQHGGPQFGLILALMTQGLAISDYPDMVRVSTLAESYG-FDS
MUL_3202|M.ulcerans_Agy99 MHENTSNPPQHGGPQFGLILALMTQGLAISDYPDMVRVSTLAESYG-FDS
MAV_0169|M.avium_104 ------------------MRFAFKTSPQNTTWAQMLAVWQAADDIDVYES
MSMEG_0702|M.smegmatis_MC2_155 -----------------MDFRVFVEPQQGATYSDQLAVAQAAERLG-YSA
TH_2824|M.thermoresistible__bu -----------------MDFRVFVEPQQGGTYADQLAVARAAEDLG-YSA
Mflv_0268|M.gilvum_PYR-GCK --------MSSV------DFRVFVEPQQGASYADQLAVARTAEQAG-YSA
Mvan_0629|M.vanbaalenii_PYR-1 --------MSTVAATNTPDFRIFVEPQQGASYADQLAVARTAESAG-YSA
MAB_4277|M.abscessus_ATCC_1997 -----------------MDFRVFVEPQQGATYSDQLRVAQAAEELG-FSA
MLBr_00348|M.leprae_Br4923 ----------MVERANAMKLGLQLGYWAAQPPGNAAELVAAAEGAG-FDA
: : *: . :.:
Mb0815c|M.bovis_AF2122/97 VWLCDHFLTISPGEYAKVAGIAADTGSATGTETGGAGQCAPSRSLPLLEC
Rv0791c|M.tuberculosis_H37Rv VWLCDHFLTISPGEYAKVAGIAADTGSATGTETGGAGQCAPSRSLPLLEC
MMAR_2559|M.marinum_M VWLCDHFLTLSPDDYVTQAGISGEHERRNGRDR-------PA-TMPLLEA
MUL_3202|M.ulcerans_Agy99 VWLCDHFLTLSPDDYVTQAGISGEHERRDGRDR-------PG-TMPLLEA
MAV_0169|M.avium_104 GWTFDHFYPIFSDS-----------------------------SGPCLEG
MSMEG_0702|M.smegmatis_MC2_155 FFRSDHYLKMAG-DGLPG----------------------------PTDS
TH_2824|M.thermoresistible__bu FFRSDHYLAMSG-DGLPG----------------------------PTDS
Mflv_0268|M.gilvum_PYR-GCK FFRSDHYLAMAG-DGLPG----------------------------PTDS
Mvan_0629|M.vanbaalenii_PYR-1 FFRSDHYVAMAG-DGLPG----------------------------PTDS
MAB_4277|M.abscessus_ATCC_1997 FFRSDHYLAMGGADGLPG----------------------------PTDA
MLBr_00348|M.leprae_Br4923 VFTGEAWG---------------------------------------SDA
: : : :
Mb0815c|M.bovis_AF2122/97 WTALAALSRDTTKLRLGTSVLCNSYRHPSVLAKMAATLDVISQGRLDLGL
Rv0791c|M.tuberculosis_H37Rv WTALAALSRDTTKLRLGTSVLCNSYRHPSVLAKMAATLDVISQGRLDLGL
MMAR_2559|M.marinum_M WTALSALARDTTYIRLGTSVLCNSYRHPAVLAKMAATLDIISEGRLDLGL
MUL_3202|M.ulcerans_Agy99 WTALSALARDTTYIRLGTSVLCNSYRHPAVLAKMAATLDIISEGRLDLGL
MAV_0169|M.avium_104 WTTLTALAQATKRLRLGTLVTGIHYRHPAVLANMAAAVDIISGGRLELGI
MSMEG_0702|M.smegmatis_MC2_155 WVTLAGIARETSTIRLGTMVTSATFRYPGPLAISVAQVDEMSGGRVELGL
TH_2824|M.thermoresistible__bu WVTLAGIARETTSIRLGTLVTSATFRYPGPLAISVAQVDEMSGGRVEFGL
Mflv_0268|M.gilvum_PYR-GCK WVTLAGLARETSSIRLGTMVTSATFRHPGPLAISVAQVDEMSGGRVELGL
Mvan_0629|M.vanbaalenii_PYR-1 WVTLAGIARETSTIRLGTMVTSATFRYPGPLAISVAQVDEMSGGRVEFGI
MAB_4277|M.abscessus_ATCC_1997 WVTLGAIARETTAIRLGTLVTSATFRHPGPLAVTVAQVDEMSSGRVDFGL
MLBr_00348|M.leprae_Br4923 YTPLAWWGSSTQRVRLGTSVVQLSARTPTACAMAALTLDHLSGGRHILGL
:..* . * :**** * * * * . :* :* ** :*:
Mb0815c|M.bovis_AF2122/97 GAGWFRRESQAYGIPFPPVGDRVSALAESLQVIKAVWTEP---NPTYAGR
Rv0791c|M.tuberculosis_H37Rv GAGWFRRESQAYGIPFPPVGDRVSALAESLQVIKAVWTEP---NPTYAGR
MMAR_2559|M.marinum_M GAGWFEREFTAYGIPFLPVGDRVTGLGEALEVITAIWTQP---DPVYSGR
MUL_3202|M.ulcerans_Agy99 GAGWFEREFTAYGIPFLPVGDRVTGLGEALEVITAIWTQP---DPVYSGR
MAV_0169|M.avium_104 GAGWNEEESGAYGIELGSIRERFDRFEEACAVLTSLLSQE---TTDFDGK
MSMEG_0702|M.smegmatis_MC2_155 GAGWFEEEHLAYAIPFPPLGERFDRLTEQLQILTGMWGTPVGDTFDFAGT
TH_2824|M.thermoresistible__bu GSGWFEPEHQAYAIPFPPLGERFDRLTEQLEIITGLWTTPSGEKYDFTGK
Mflv_0268|M.gilvum_PYR-GCK GAGWFEAEHKAYAIPFPPLGERFDRLTEQLEILTGLWTTPTGETFDYSGT
Mvan_0629|M.vanbaalenii_PYR-1 GAGWFEAEHKAYAIPFPPLGERFDRLTEQLEILTGLWTAAPGETFDYSGR
MAB_4277|M.abscessus_ATCC_1997 GAGWFSEEHEAYAIPFPSLGERFDRLEEQLEILTGLWTTPTGDTYSYAGK
MLBr_00348|M.leprae_Br4923 GVSGPQVVEGWYGQPFP---KPLSRTREYIDIVRQVWAREAPVVSDGQHY
* . *. : . . * :: :
Mb0815c|M.bovis_AF2122/97 FYTLDGATCD-----PPPVQRPH---PPLWIGGEGD-RVQRIAAKHAQGL
Rv0791c|M.tuberculosis_H37Rv FYTLDGATCD-----PPPVQRPH---PPLWIGGEGD-RVQRIAAKHAQGL
MMAR_2559|M.marinum_M FYTLDGAICD-----PPPVQTPH---PPLWVGGEGD-RVHRIAARSAQGI
MUL_3202|M.ulcerans_Agy99 FYTLDGAICD-----PPPVQTPH---PPLWVGGEGD-RVHRIAARSAQGI
MAV_0169|M.avium_104 FYQLKGARNE-----PKGPQRPH---PPICIGGSGEKRTLRITARYAQHW
MSMEG_0702|M.smegmatis_MC2_155 HYTVKDSPGL-----PKPVQTPH---PPIIIGGQGAKRTPALAAQFASEF
TH_2824|M.thermoresistible__bu HYTVVDSPGL-----PKPVQDPY---PPIIIGGQGPKRTPALAARFANEF
Mflv_0268|M.gilvum_PYR-GCK HYSIADSPAL-----PKPAQSPR---PPIVIGGRGAKRTPALTARFADEF
Mvan_0629|M.vanbaalenii_PYR-1 HYSIVDSPAL-----PKPAQRPH---PPIIIGGGGAKRTPALAARFADEF
MAB_4277|M.abscessus_ATCC_1997 HYTVTDSPAL-----PKPVQEPH---PPIVIGGLGVKRTPALAARFATEF
MLBr_00348|M.leprae_Br4923 PLPLTGAGATGLGKALKPITHPLRADIPIMLGAEGP-KNVALAAEICDGW
: .: * *: :*. * : ::*. .
Mb0815c|M.bovis_AF2122/97 NVRWWSPQQVTQRRGFLTQASEAAGR----------DPDTLRLSVTLLLA
Rv0791c|M.tuberculosis_H37Rv NVRWWSPQQVTQRRGFLTQASEAAGR----------DPDTLRLSVTLLLA
MMAR_2559|M.marinum_M NVRWWSPQKVKQRKDFLAQACATAQR----------DPTTLRFSVTALLA
MUL_3202|M.ulcerans_Agy99 NVRWWLPQKVKQRKDFLAQTCATAQR----------DPTTLRFSVTALLA
MAV_0169|M.avium_104 NFVGGPPDVFAHKRDVLASHCADIGR----------DPKEILLSAHLRLE
MSMEG_0702|M.smegmatis_MC2_155 NIPFVPLDTLKTQFERVAAAVEGAGR----------SADSMIYSAAFVLC
TH_2824|M.thermoresistible__bu NIPFVRLDALKAQFERVAAAVAEAGR----------EPDSMIYSAAFVLC
Mflv_0268|M.gilvum_PYR-GCK NIPFVPLDELTTQYARVAAAVEDAGR----------AKDSLTYSAAFVVC
Mvan_0629|M.vanbaalenii_PYR-1 NIPFVPLDTLTEQYGRVAAAVEQAGR----------AKDSLIYSAAFVLC
MAB_4277|M.abscessus_ATCC_1997 NLPFQPVDVITDQYARVAHAVEAAGR----------PADSMTYSAAFALC
MLBr_00348|M.leprae_Br4923 LPIFFSPRMADMYNEWLDEGFARTGARRSREDFEICASAQIVVTEDRAAA
: : :
Mb0815c|M.bovis_AF2122/97 P----TQSGEEEVRIREEFASIPEPGLIVGTPDRCVERIREYQDRGVGHF
Rv0791c|M.tuberculosis_H37Rv P----TQSGEEEVRIREEFASIPEPGLIVGTPDRCVERIREYQDRGVGHF
MMAR_2559|M.marinum_M P----TDSAEQEAQIRTEFSAVPYDGLIVGPADRCIERINEYYESGVDTF
MUL_3202|M.ulcerans_Agy99 P----TDSAEQEAQIRTEFSAVPYDGLIVGPADRCIERINEYYESGVDTF
MAV_0169|M.avium_104 P----DLDYG---KVIDDAAALAAEGLDLG--------IVYLPPPHDPAV
MSMEG_0702|M.smegmatis_MC2_155 AGADEAQIAKRAAAIGREVDELRSNSPLVGTSTEIVDKLGPFIEAGVQRV
TH_2824|M.thermoresistible__bu AGRDDAEIARRAAAINREVDELRSNSPVVGTPAEIVDRLGPFIEAGVQRV
Mflv_0268|M.gilvum_PYR-GCK VGRDDAEIARRAAAIGREVDEMRSNSPTVGTPAEVVDKLGPFLEAGVERV
Mvan_0629|M.vanbaalenii_PYR-1 VGRDETEIARRAAAIGREVDELRSNSPVVGTPGEVVDKLRPFLEAGVQRI
MAB_4277|M.abscessus_ATCC_1997 IGDTDADIARRAANIGREVEEITENSPLAGTPDAVAEKLSIYVDAGVQRV
MLBr_00348|M.leprae_Br4923 FAGIKPFLALYMGGMGAEGTNFHADVYRRMGYAEVVDEVTALFRSNQKDK
: : . :
Mb0815c|M.bovis_AF2122/97 LFTIPHVVKSD-----------------------YLHIIGSDIIPRVKTE
Rv0791c|M.tuberculosis_H37Rv LFTIPHVVKSD-----------------------YLHIIGSDIIPRVKTE
MMAR_2559|M.marinum_M LFTIPDVARSD-----------------------LIHTVGQEVLPAFD--
MUL_3202|M.ulcerans_Agy99 LFTIPDVARSD-----------------------LIHTVGQEVLPAFD--
MAV_0169|M.avium_104 LEPLAEAIRDS-----------------------GLLRS-----------
MSMEG_0702|M.smegmatis_MC2_155 YLQVLDMSDLD-----------------------HVEFFANEVVRQFS--
TH_2824|M.thermoresistible__bu YLQLLDMSDLD-----------------------HLELIATQVVPQLR--
Mflv_0268|M.gilvum_PYR-GCK YLQVLDMSDLE-----------------------HVEFFAEQVIPQIH--
Mvan_0629|M.vanbaalenii_PYR-1 YLQLLDMSDLD-----------------------HVQFFAEQVIPRIG--
MAB_4277|M.abscessus_ATCC_1997 YVQLLDIRDLD-----------------------HLEFFASTVIPQFS--
MLBr_00348|M.leprae_Br4923 AAEIIPNELVDSTMIVGDVDYVCKQIAAWSAAGVTMMMVSASSVEQVRDL
: . :
Mb0815c|M.bovis_AF2122/97 VTIP
Rv0791c|M.tuberculosis_H37Rv VTIP
MMAR_2559|M.marinum_M ----
MUL_3202|M.ulcerans_Agy99 ----
MAV_0169|M.avium_104 ----
MSMEG_0702|M.smegmatis_MC2_155 ----
TH_2824|M.thermoresistible__bu ----
Mflv_0268|M.gilvum_PYR-GCK ----
Mvan_0629|M.vanbaalenii_PYR-1 ----
MAB_4277|M.abscessus_ATCC_1997 ----
MLBr_00348|M.leprae_Br4923 AGLF