For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
SRAVVVVPAHNEKDHLPDCLRALTTAALCVPIPVGIVVVLDSCDDGSDALAGQFGADVHFVSVDAGNVGA TRAAGFEYARTLSGDDEIGRTWYATTDADSTVDADWLVRMLGAGADMVLGVVRVSNWRHFSPAVARRYLR GYRSRGTGHRHIHGANMGFRAESYWRVGGFRALKSSEDVDLVRRFEDAGMFIHRDRRLSVATSDRREGRA PGGFAQHLRNLSRSDDPGKADVA*
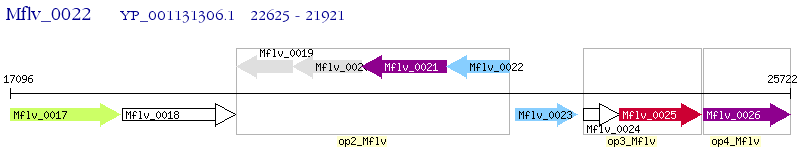
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0022 | - | - | 100% (234) | glycosyl transferase family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2077c | ppm1 | 3e-06 | 32.39% (176) | polyprenol-monophosphomannose synthase Ppm1 |
| M. tuberculosis H37Rv | Rv2051c | ppm1 | 3e-06 | 32.39% (176) | polyprenol-monophosphomannose synthase Ppm1 |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2207 | - | 9e-05 | 32.31% (130) | polyprenol phosphate mannosyl transferase 1 |
| M. marinum M | - | - | - | - | - |
| M. avium 104 | - | - | - | - | - |
| M. smegmatis MC2 155 | MSMEG_1097 | - | 4e-70 | 57.69% (234) | glycosyl transferase, group 2 family protein |
| M. thermoresistible (build 8) | - | - | - | - | - |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_3418 | - | 2e-05 | 31.43% (175) | dolichyl-phosphate beta-D-mannosyltransferase |
CLUSTAL 2.0.9 multiple sequence alignment
MAB_2207|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_3418|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2077c|M.bovis_AF2122/97 MKLGAWVAAQLPTTRTAVRTRLTRLVVSIVAGLLLYASFPPRNCWWAAVV
Rv2051c|M.tuberculosis_H37Rv MKLGAWVAAQLPTTRTAVRTRLTRLVVSIVAGLLLYASFPPRNCWWAAVV
Mflv_0022|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_1097|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2207|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_3418|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2077c|M.bovis_AF2122/97 ALALLAWVLTHRATTPVGGLGYGLLFGLVFYVSLLPWIGELVGPGPWLAL
Rv2051c|M.tuberculosis_H37Rv ALALLAWVLTHRATTPVGGLGYGLLFGLVFYVSLLPWIGELVGPGPWLAL
Mflv_0022|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_1097|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2207|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_3418|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2077c|M.bovis_AF2122/97 ATTCALFPGIFGLFAVVVRLLPGWPIWFAVGWAAQEWLKSILPFGGFPWG
Rv2051c|M.tuberculosis_H37Rv ATTCALFPGIFGLFAVVVRLLPGWPIWFAVGWAAQEWLKSILPFGGFPWG
Mflv_0022|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_1097|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2207|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_3418|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2077c|M.bovis_AF2122/97 SVAFGQAEGPLLPLVQLGGVALLSTGVALVGCGLTAIALEIEKWWRTGGQ
Rv2051c|M.tuberculosis_H37Rv SVAFGQAEGPLLPLVQLGGVALLSTGVALVGCGLTAIALEIEKWWRTGGQ
Mflv_0022|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_1097|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2207|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_3418|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2077c|M.bovis_AF2122/97 GDAPPAVVLPAACICLVLFAAIVVWPQVRHAGSGSGGEPTVTVAVVQGNV
Rv2051c|M.tuberculosis_H37Rv GDAPPAVVLPAACICLVLFAAIVVWPQVRHAGSGSGGEPTVTVAVVQGNV
Mflv_0022|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_1097|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2207|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_3418|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2077c|M.bovis_AF2122/97 PRLGLDFNAQRRAVLDNHVEETLRLAADVHAGLAQQPQFVIWPENSSDID
Rv2051c|M.tuberculosis_H37Rv PRLGLDFNAQRRAVLDNHVEETLRLAADVHAGLAQQPQFVIWPENSSDID
Mflv_0022|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_1097|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2207|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_3418|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2077c|M.bovis_AF2122/97 PFVNPDAGQRISAAAEAIGAPILIGTLMDVPGRPRENPEWTNTAIVWNPG
Rv2051c|M.tuberculosis_H37Rv PFVNPDAGQRISAAAEAIGAPILIGTLMDVPGRPRENPEWTNTAIVWNPG
Mflv_0022|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_1097|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2207|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_3418|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2077c|M.bovis_AF2122/97 TGPADRHDKAIVQPFGEYLPMPWLFRHLSGYADRAGHFVPGNGTGVVRIA
Rv2051c|M.tuberculosis_H37Rv TGPADRHDKAIVQPFGEYLPMPWLFRHLSGYADRAGHFVPGNGTGVVRIA
Mflv_0022|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_1097|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2207|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_3418|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2077c|M.bovis_AF2122/97 GVPVGVATCWEVIFDRAPRKSILGGAQLLTVPSNNATFNKTMSEQQLAFA
Rv2051c|M.tuberculosis_H37Rv GVPVGVATCWEVIFDRAPRKSILGGAQLLTVPSNNATFNKTMSEQQLAFA
Mflv_0022|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_1097|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2207|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_3418|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2077c|M.bovis_AF2122/97 KVRAVEHDRYVVVAGTTGISAVIAPDGGELIRTDFFQPAYLDSQVRLKTR
Rv2051c|M.tuberculosis_H37Rv KVRAVEHDRYVVVAGTTGISAVIAPDGGELIRTDFFQPAYLDSQVRLKTR
Mflv_0022|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_1097|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2207|M.abscessus_ATCC_1997 --------------------------------------------------
Mvan_3418|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb2077c|M.bovis_AF2122/97 LTPATRWGPILQWILVGAAAAVVLVAMRQNGWFPRPRRSEPKGENDDSDA
Rv2051c|M.tuberculosis_H37Rv LTPATRWGPILQWILVGAAAAVVLVAMRQNGWFPRPRRSEPKGENDDSDA
Mflv_0022|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_1097|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2207|M.abscessus_ATCC_1997 -------------------------------------MDP---VTE----
Mvan_3418|M.vanbaalenii_PYR-1 -------------------------------------MSVPGGMTDGP--
Mb2077c|M.bovis_AF2122/97 PPGRSEASGPPALSESDDELIQPEQGGRHSSGFGRHRATSRSYMTTGQPA
Rv2051c|M.tuberculosis_H37Rv PPGRSEASGPPALSESDDELIQPEQGGRHSSGFGRHRATSRSYMTTGQPA
Mflv_0022|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_1097|M.smegmatis_MC2_155 --------------------------------------------------
MAB_2207|M.abscessus_ATCC_1997 ------RPSARTLVIIPTYNERENLPLILGRLHKAQPDVHVLIVDDGSPD
Mvan_3418|M.vanbaalenii_PYR-1 --GPGTRPSDRTLVIIPTYNERQNLPLIVGRVHQARPDVDILIVDDDSPD
Mb2077c|M.bovis_AF2122/97 PPAPGNRPSQRVLVIIPTFNERENLPVIHRRLTQACPAVHVLVVDDSSPD
Rv2051c|M.tuberculosis_H37Rv PPAPGNRPSQRVLVIIPTFNERENLPVIHRRLTQACPAVHVLVVDDSSPD
Mflv_0022|M.gilvum_PYR-GCK --------MSRAVVVVPAHNEKDHLPDCLRALTTAALCVPIPVGIVVVLD
MSMEG_1097|M.smegmatis_MC2_155 ---MLESQVDQIAVVIPAHNERAHLPMSLRAMVTAAMCVPVPVQIVVVLD
: *::*:.**: :** : * * : : *
MAB_2207|M.abscessus_ATCC_1997 GTGELADEAALADPDRVHVMHRKEKGGLGAAYIAGFGWGLAR------QY
Mvan_3418|M.vanbaalenii_PYR-1 GTGQLADELSVADPDRIHVMHRTAKEGLGAAYLAGFAWGLGR------QY
Mb2077c|M.bovis_AF2122/97 GTGQLADELAQADPGRTHVMHRTAKNGLGAAYLAGFAWGLSR------EY
Rv2051c|M.tuberculosis_H37Rv GTGQLADELAQADPGRTHVMHRTAKNGLGAAYLAGFAWGLSR------EY
Mflv_0022|M.gilvum_PYR-GCK SCDDGSDALAGQFGADVHFVSVDAGN-VGATRAAGFEYARTLSGDDEIGR
MSMEG_1097|M.smegmatis_MC2_155 ACTDGSEALAGDFGSDVHFVSVDAGN-VGAARAAGFAYARSLRPDT--VR
. : :: : *.: :**: *** :.
MAB_2207|M.abscessus_ATCC_1997 SVLVEMDADGSHAPEQLSRLLD-AVDAGADLAIGSRYVPGGTVVNWPWRR
Mvan_3418|M.vanbaalenii_PYR-1 TVLVEMDADGSHPPEQLARLLD-AVDAGADVVIGSRYVSGGEVVNWPRRR
Mb2077c|M.bovis_AF2122/97 SVLVEMDADGSHAPEQLQRLLD-AVDAGADLAIGSRYVAGGTVRNWPWRR
Rv2051c|M.tuberculosis_H37Rv SVLVEMDADGSHAPEQLQRLLD-AVDAGADLAIGSRYVAGGTVRNWPWRR
Mflv_0022|M.gilvum_PYR-GCK TWYATTDADSTVDADWLVRMLG----AGADMVLGVVRVSNWRHFSPAVAR
MSMEG_1097|M.smegmatis_MC2_155 TWYATSDADTAVPADWLVEMTTTAVEVDADMVLGGVHVPGGQDFS-----
: . *** : .: * .: ..**:.:* *.. .
MAB_2207|M.abscessus_ATCC_1997 LVLSRSANVYARLVLGVKPHDITAGYRAYRREVLEKLDLGAVESHGYCFQ
Mvan_3418|M.vanbaalenii_PYR-1 LVLSRTANGYSRILLGVDIHDITAGYRAYRREVLEKIDLAAVDTKGYGFQ
Mb2077c|M.bovis_AF2122/97 LVLSKTANTYSRLALGIGIHDITAGYRAYRREALEAIDLDGVDSKGYCFQ
Rv2051c|M.tuberculosis_H37Rv LVLSKTANTYSRLALGIGIHDITAGYRAYRREALEAIDLDGVDSKGYCFQ
Mflv_0022|M.gilvum_PYR-GCK RYLRGYR---SRGTGHRHIHGANMGFRAES-----YWRVGGFRALKSSED
MSMEG_1097|M.smegmatis_MC2_155 -------------ESNDHVHGANMGFRADA-----YWSVGGFRGLKTGED
*. . *:** : .. :
MAB_2207|M.abscessus_ATCC_1997 IDLTLRTIAHGFEVAEVPITFTERAIGESKMSGSIINEALVKVTKWGVQS
Mvan_3418|M.vanbaalenii_PYR-1 VDLTWRSINAGFTVIEVPITFTEREHGVSKMDGSTIREAIIKVAQWGLRA
Mb2077c|M.bovis_AF2122/97 IDLTWRTVSNGFVVTEVPITFTERELGVSKMSGSNIREALVKVARWGIEG
Rv2051c|M.tuberculosis_H37Rv IDLTWRTVSNGFVVTEVPITFTERELGVSKMSGSNIREALVKVARWGIEG
Mflv_0022|M.gilvum_PYR-GCK VDLVRRFEDAGMFIHRDRRLSVATSDRREGRAPGGFAQHLRNLSRSDDPG
MSMEG_1097|M.smegmatis_MC2_155 AELADRFESHGLHVHRDGDLSVETSARRSGRAPDGFADYLRELARTPVPA
:*. * *: : . . . . : : : :::: .
MAB_2207|M.abscessus_ATCC_1997 RLDR---ARG-------------VNR-----
Mvan_3418|M.vanbaalenii_PYR-1 RLDR---ARG-------------TTR-----
Mb2077c|M.bovis_AF2122/97 RLSRSDHARARPDIARPGAGGSRVSRADVTE
Rv2051c|M.tuberculosis_H37Rv RLSRSDHARARPDIARPGAGGSRVSRADVTE
Mflv_0022|M.gilvum_PYR-GCK KADVA--------------------------
MSMEG_1097|M.smegmatis_MC2_155 ELGAPA-------------------------
. .