For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTEAPDVDLADGNFYASREARAAYRWMRANQPVFRDRNGLAAASTYQAVIDAERQPELFSNAGGIRPDQP ALPMMIDMDDPAHLLRRKLVNAGFTRKRVKDKEASIAALCDTLIDAVCERGECDFVRDLAAPLPMAVIGD MLGVRPEQRDMFLRWSDDLVTFLSSHVSQEDFQITMDAFAAYNDFTRATIAARRADPPTTWSACW
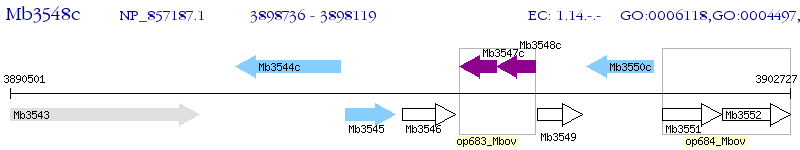
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3548c | cyp142a | - | 100% (205) | cytochrome P450 monooxygenase |
| M. bovis AF2122 / 97 | Mb2289 | cyp124 | 9e-15 | 28.71% (202) | cytochrome P450 124 CYP124 |
| M. bovis AF2122 / 97 | Mb0789c | cyp123 | 2e-14 | 32.84% (134) | cytochrome P450 123 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1541 | - | 8e-83 | 78.24% (193) | cytochrome P450 |
| M. tuberculosis H37Rv | Rv3518c | cyp142 | 1e-112 | 100.00% (197) | cytochrome P450 monooxygenase 142 |
| M. leprae Br4923 | MLBr_02088 | - | 6e-11 | 32.62% (141) | putative cytochrome p450 |
| M. abscessus ATCC 19977 | MAB_1216c | - | 4e-14 | 29.94% (177) | cytochrome P450 |
| M. marinum M | MMAR_5003 | cyp142A3 | 1e-104 | 94.30% (193) | cytochrome P450 142A3 Cyp142A3 |
| M. avium 104 | MAV_0641 | - | 2e-83 | 76.92% (195) | P450 heme-thiolate protein |
| M. smegmatis MC2 155 | MSMEG_5918 | - | 2e-83 | 75.13% (197) | P450 heme-thiolate protein |
| M. thermoresistible (build 8) | TH_3500 | - | 1e-79 | 72.31% (195) | PUTATIVE cytochrome P450 |
| M. ulcerans Agy99 | MUL_4077 | cyp142A3 | 1e-102 | 93.26% (193) | cytochrome P450 142A3 Cyp142A3 |
| M. vanbaalenii PYR-1 | Mvan_5217 | - | 3e-83 | 77.44% (195) | cytochrome P450 |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3548c|M.bovis_AF2122/97 ------------------------MTE-----APDVDLADGNFYASR-EA
Rv3518c|M.tuberculosis_H37Rv ------------------------MTE-----APDVDLADGNFYASR-EA
MMAR_5003|M.marinum_M ------------------------MTKPLI--KPDVDLTDGNFYASR-QA
MUL_4077|M.ulcerans_Agy99 ----------------------MAMTKPLI--KPDVDLTDGNFYASR-QA
Mflv_1541|M.gilvum_PYR-GCK ------------------------MTSTVET-GLDVDLADGNFYADGTAA
Mvan_5217|M.vanbaalenii_PYR-1 ------------------------MTSTLP--ALDVDLADGNFYADGRAA
MSMEG_5918|M.smegmatis_MC2_155 ------------------------MTQMLT--RPDVDLVNGMFYADG-GA
MAV_0641|M.avium_104 ------------------------MTSTIPEAIANIDLADGNFYADRRAS
TH_3500|M.thermoresistible__bu ------------------------MTST----APDIDLTDGRFYADSASA
MLBr_02088|M.leprae_Br4923 MRTCVPTRTCVYAFIEYLSHNRPMGTNPPSLVEAQMLLLRLIDPGTRADP
MAB_1216c|M.abscessus_ATCC_199 --------------------------MTVAETSAVPLVFDPYDYDFHEDP
: .
Mb3548c|M.bovis_AF2122/97 RAAYRWMRANQPVFRDR-NGLAAASTYQAVIDAERQPELFSNAGGIRPDQ
Rv3518c|M.tuberculosis_H37Rv RAAYRWMRANQPVFRDR-NGLAAASTYQAVIDAERQPELFSNAGGIRPDQ
MMAR_5003|M.marinum_M REAYRWMRANQPVFRDR-NGLAAASTYQAVIDAERQPELFSNAGGIRPDQ
MUL_4077|M.ulcerans_Agy99 REAYRWMRANQPVFRDR-NGLAAASTYQAVIDAERQPELFSNAGGIRPDQ
Mflv_1541|M.gilvum_PYR-GCK RAAYAWMRANQPVFRDR-NGLAAATTYQAVLDAERNPELFSSTGGIRPDQ
Mvan_5217|M.vanbaalenii_PYR-1 REAYKWMRANQPVFRDR-NGLAAATTYQAVLDAERNPELFSSTGGIRPDQ
MSMEG_5918|M.smegmatis_MC2_155 REAYRWMRANEPVFRDR-NGLAAATTYQAVLDAERNPELFSSTGGIRPDQ
MAV_0641|M.avium_104 REAYRWMRANQPVFRDR-NGLAGATTYQAIQDAERNPELFSSTGGIRPDQ
TH_3500|M.thermoresistible__bu RAAYRWMRQNQPVFRDR-NGLAAAATYQAIIESERNPELFSNTGGIRPEH
MLBr_02088|M.leprae_Br4923 FPVYRALIDYGPMQLPG-MPLTVFSSFSDCDEALRHPLSASDRLKATLAQ
MAB_1216c|M.abscessus_ATCC_199 YPYYRRLRDEAPLYRNDDLKFWALSRHHDVLQGFRNSEALSNANGVSMDK
* : *: : : . :. *:. *. :
Mb3548c|M.bovis_AF2122/97 PALP-----------MMIDMDDPAHLLRRKLVNAGFTRKRVKDKEASIAA
Rv3518c|M.tuberculosis_H37Rv PALP-----------MMIDMDDPAHLLRRKLVNAGFTRKRVKDKEASIAA
MMAR_5003|M.marinum_M DALP-----------MMIDMDDPAHLLRRKLVNAGFTRKRVKDKERSIAQ
MUL_4077|M.ulcerans_Agy99 DALP-----------MMIDMDDPAHLWRRKLVNAGFTRKRVKDKEHSIAQ
Mflv_1541|M.gilvum_PYR-GCK PGMP-----------YMIDMDDPAHLLRRKLVNSGFTRKRVMDKVPSIEK
Mvan_5217|M.vanbaalenii_PYR-1 PGMP-----------YMIDMDDPAHLLRRKLVNSGFTRKRVMDKVPSIEN
MSMEG_5918|M.smegmatis_MC2_155 PGMP-----------YMIDMDDPQHLLRRKLVNAGFTRKRVMDKVDSIGR
MAV_0641|M.avium_104 PGMP-----------YMIDMDDPAHLLRRKLVNAGFTRKRVKEKEPSIGT
TH_3500|M.thermoresistible__bu PGMP-----------YMIDMDDPEHLVRRKLVNHGFTRKRVRDREDSIAR
MLBr_02088|M.leprae_Br4923 QAIAAGAEPRPFYASSFMFLDPPDHTRLRKLVSKAFAPKVVQALEGDIAA
MAB_1216c|M.abscessus_ATCC_199 ASFGPHAK----LVMSFLAMDDPEHLRLRALVSKGFTPRRIRELEGQVVA
.: :: :* * * * **. .*: : : .:
Mb3548c|M.bovis_AF2122/97 LCDTLIDAVCERG---ECDFVRDLAAPLPMAVIGDMLGVRPEQRDMFLRW
Rv3518c|M.tuberculosis_H37Rv LCDTLIDAVCERG---ECDFVRDLAAPLPMAVIGDMLGVRPEQRDMFLRW
MMAR_5003|M.marinum_M LCDTLIDAVCERG---ECDFVRDLAAPLPMAVIGDMLGVLPEQREMFLRW
MUL_4077|M.ulcerans_Agy99 LCDTLIDAVCERG---ECDFVRDLAAPLPMAVIGDMLGVLPEQREMFLRW
Mflv_1541|M.gilvum_PYR-GCK LCDTLIDAVCERG---ECDFVRDIAAPLPMAVIGDMLGVLPEERDMLLTW
Mvan_5217|M.vanbaalenii_PYR-1 LCDTLIDAVCERG---ECDFVRDIAAPLPMAVIGDMLGVLPDERDMLLKW
MSMEG_5918|M.smegmatis_MC2_155 LCDTLIDAVCERG---ECDFVRDIAAPLPMAVIGDMLGVLPTERDMLLKW
MAV_0641|M.avium_104 LCDTLIDAVCERG---ECDFVRDIAAPLPMAVIGDMLGVLPTERGMLLKW
TH_3500|M.thermoresistible__bu LCDTLIDNVCERG---ECDFVRDIAAPLPMAVIGDMLGVAPEDRGQLLKW
MLBr_02088|M.leprae_Br4923 LVDSLLDKGAAAG---QFDVIADLAFPLAVAVICRLLGVPYEDAPEFGRV
MAB_1216c|M.abscessus_ATCC_199 LARTHLDRALANASDRSFDFIAEYAGKLPMDVISELMGVPESDRARIREL
* : :* . . *.: : * *.: ** ::** : :
Mb3548c|M.bovis_AF2122/97 SDDLVTFLSSHVS-----QEDFQITMDAFAAYNDFTRATIAARRADPPTT
Rv3518c|M.tuberculosis_H37Rv SDDLVTFLSSHVS-----QEDFQITMDAFAAYNDFTRATIAARRADPTDD
MMAR_5003|M.marinum_M SDDLVTFLSSHVS-----QEDFQVTIDAFAAYNDFTRATIAARRAEPTDD
MUL_4077|M.ulcerans_Agy99 SDDLVTFLSSQVS-----QEDFQVTIDAFAAYNDFTRATIAARRAEPTDD
Mflv_1541|M.gilvum_PYR-GCK SDDLVCGLSSTVD-----EQTIQKLMDTFAAYTAFTMEVIADRRANPRDD
Mvan_5217|M.vanbaalenii_PYR-1 SDDLVCGLSSTVD-----EQTIQKLMDTFAAYTAFTTEVIADRRANPRDD
MSMEG_5918|M.smegmatis_MC2_155 SDDLVCGLSSHVD-----EAAIQKLMDTFAAYTEFTKDVITKRRAEPTDD
MAV_0641|M.avium_104 SDDLVCGLSSHIDPT---SAEFQTVMDAFAAYTAFTMDIIAKRRAEPTDD
TH_3500|M.thermoresistible__bu SDDLVTGLSTTAD-----EATVQAVMDAFAGYTAYTMEIIGKRRAEPTDD
MLBr_02088|M.leprae_Br4923 SALLVQSVDPFITITGEPPEATEERLRAGVWLRDYLEQLVKCRRGTPGED
MAB_1216c|M.abscessus_ATCC_199 ADGVMHREDGLADVP-------PEAIQASFDLMTYYIEMVRERRRRPTED
: :: . : : : : ** *
Mb3548c|M.bovis_AF2122/97 WSACW---------------------------------------------
Rv3518c|M.tuberculosis_H37Rv LVSVLVSSEVDGERLSDDELVMETLLILIGGDETTRHTLSGGTEQLLRNR
MMAR_5003|M.marinum_M LVSVLVSSEVDGERLSDDELVMETLLILIGGDETTRHTLSGGSEQLLRNR
MUL_4077|M.ulcerans_Agy99 LVSVLVSSEVDGERLSDDELVMETLLILIGGDETTRHTLSGGSEQLLRNR
Mflv_1541|M.gilvum_PYR-GCK LFSILVNAEVEGSRMSDDEIVFETLLILIGGDETTRHTLSGGTEQILRHQ
Mvan_5217|M.vanbaalenii_PYR-1 LFSVLVNAEVEGQRMSDDEIVFETLLILIGGDETTRHTLSGGTEQLLRHQ
MSMEG_5918|M.smegmatis_MC2_155 LFSVLVNSEVEGQRMSDDEIVFETLLILIGGDETTRHTLSGGTEQLLRHR
MAV_0641|M.avium_104 LFSILVNAEVEGQRMSDDEIVMETLLILIGGDETTRHTLSGGTEQLLRHR
TH_3500|M.thermoresistible__bu LFSILINAEVEGQRMSDEEIVFETLLILIGGDETTRHTLSGGVEQLLRHP
MLBr_02088|M.leprae_Br4923 LISRLIELDESGDQLTEEEIIATCGLLLVAGHETTVNLIANAVLAMLRNP
MAB_1216c|M.abscessus_ATCC_199 LTSALLQAEIDGDRLTDEEVLAFLFLMVIAGNETTTKLLANAVYWGHRNP
:
Mb3548c|M.bovis_AF2122/97 --------------------------------------------------
Rv3518c|M.tuberculosis_H37Rv DQWDLLQRDPSLLPGAIEEMLRWTAPVKNMCRVLTADTEFHGTALCAGEK
MMAR_5003|M.marinum_M DQWDLLQSDRELLPGAIEEMLRWTAPVKNMCRMLTADTEFHGTALSEGEK
MUL_4077|M.ulcerans_Agy99 VQWDLLQSDRELLPGAIEEMLRWTAPVKNMCRMLTADTEFHGTALSKGEK
Mflv_1541|M.gilvum_PYR-GCK DQWRQVVAQPDLLPSAIEEMLRWTSPVKNMCRTLTADTEFHGTALKSGEK
Mvan_5217|M.vanbaalenii_PYR-1 DQWRQAVADPELLPGAIEEMLRWTSPVKNMCRTLTADTEFHGTSLKAGEK
MSMEG_5918|M.smegmatis_MC2_155 DQWDALVADVDLLPGAIEEMLRWTSPVKNMCRTLTADTVFHGTELRAGEK
MAV_0641|M.avium_104 DQWDALVRDPSLLPGAIEEMLRWTSPVKNMCRTLTADTEFHGTELRAGEK
TH_3500|M.thermoresistible__bu DQWQRLRNDPDLVPGAIEEMLRWTSPVKNMCRTLTADTEFHGTQLRKGEK
MLBr_02088|M.leprae_Br4923 SQWKALSSNPQRAPLVVEETLRYDPAIHLIGRVAAKDMTIGQTTLTEGDT
MAB_1216c|M.abscessus_ATCC_199 DQLATVHADHDRIPLWVEESLRYDTSSQILARTVAEDITVYDTKIPAGDI
Mb3548c|M.bovis_AF2122/97 --------------------------------------------------
Rv3518c|M.tuberculosis_H37Rv MMLLFESANFDEAVFCEPEKFDVQRNP-NSHLAFGFGTHFCLGNQLARLE
MMAR_5003|M.marinum_M IMLLFESANFDEAVFTDPEKFDIQRNP-NSHLAFGFGTHFCMGNQLARLE
MUL_4077|M.ulcerans_Agy99 IMLLFESANFDEAVFTDPEKFDIQRNP-NSHLAFGFGTHFCMGNQLARLE
Mflv_1541|M.gilvum_PYR-GCK IMLMFESANFDESVFENPESFDIHRDP-NSHVAFGFGTHFCLGNQLARLE
Mvan_5217|M.vanbaalenii_PYR-1 IMLMFESANFDEAVFENPDEFDIHRNP-NSHMAFGFGTHFCLGNQLARLE
MSMEG_5918|M.smegmatis_MC2_155 IMLMFESANFDESVFGDPDNFRIDRNP-NSHVAFGFGTHFCLGNQLARLE
MAV_0641|M.avium_104 IMLLFESGNFDESVFDDPDSFDIRRNP-NSHMAFGFGTHFCLGNQLARLE
TH_3500|M.thermoresistible__bu MLLLFESANFDETVFENPDDFRIDRDP-NPHIAFGFGTHFCLGNQLARLE
MLBr_02088|M.leprae_Br4923 MVLLLAAANRDPAVYSRPDEFDPDRPS-SRHLAFAVGSHFCLGAALARLE
MAB_1216c|M.abscessus_ATCC_199 LLLLPGSANRDDRVFDDADQYRIGREIGAKLVSFGSGAHFCLGAHLARME
Mb3548c|M.bovis_AF2122/97 --------------------------------------------------
Rv3518c|M.tuberculosis_H37Rv LSLMTERVLRRLPDLRLVADDSVLPLRPANFVSGLESMPVVFTPSPPLG-
MMAR_5003|M.marinum_M LSLMTARVVQRLPDLRLADQDSRLPLRPANFVSGLESMPVVFTPSRPLS-
MUL_4077|M.ulcerans_Agy99 LSLMTARVVQRLPDLRLADQDSRLPLRPANFVSGLESMPVVFTPSRPLR-
Mflv_1541|M.gilvum_PYR-GCK LKLMLGKILQRLPDMRLAD-ENMLPLRPANFVSGLESMPVVFTPTAKVLS
Mvan_5217|M.vanbaalenii_PYR-1 LRLMLTKILQRLPDLRLAD-ESMLPLRPANFVSGLESMPVVFTPTAPVGA
MSMEG_5918|M.smegmatis_MC2_155 LRLMTERVLRRLPDLRLAD-DAPVPLRPANFVSGPESMPVVFTPSAPVLA
MAV_0641|M.avium_104 LSMMTERVLKRLPDLRLAD-DGDLPLRPANFVSGLEAMPVVFTPSAPLLR
TH_3500|M.thermoresistible__bu LTLMLRRLLQRLPDLRLAS-DDPLPLRPANFVSGPEAMPVVFTPTEPVSA
MLBr_02088|M.leprae_Br4923 ATVTLSAISARFPQVQLAG-ELVYKPNVA--MRGMSALPVQV--------
MAB_1216c|M.abscessus_ATCC_199 AKVALTELFTRISGYEIDERNSVRVHSSN--VRGFAHLPMTVQLREGR--