For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MAAVVKSVALAGRPTTPDRVHEVLGRSMLVDGLDIVLDLTRSGGSYLVDAITGRRYLDMFTFVASSALGM NPPALVDDREFHAELMQAALNKPSNSDVYSVAMARFVETFARVLGDPALPHLFFVEGGALAVENALKAAF DWKSRHNQAHGIDPALGTQVLHLRGAFHGRSGYTLSLTNTKPTITARFPKFDWPRIDAPYMRPGLDEPAM AALEAEALRQARAAFETRPHDIACFVAEPIQGEGGDRHFRPEFFAAMRELCDEFDALLIFDEVQTGCGLT GTAWAYQQLDVAPDIVAFGKKTQVCGVMAGRRVDEVADNVFAVPSRLNSTWGGNLTDMVRARRILEVIEA EGLFERAVQHGKYLRARLDELAADFPAVVLDPRGRGLMCAFSLPTTADRDELIRQLWQRAVIVLPAGADT VRFRPPLTVSTAEIDAAIAAVRSALPVVT
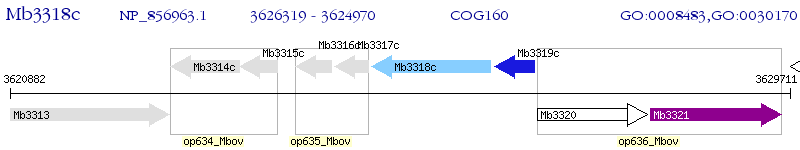
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3318c | lat | - | 100% (449) | L-lysine aminotransferase |
| M. bovis AF2122 / 97 | Mb1683 | argD | 3e-26 | 28.92% (408) | acetylornithine aminotransferase |
| M. bovis AF2122 / 97 | Mb2620 | gabT | 9e-23 | 27.62% (420) | 4-aminobutyrate aminotransferase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_4796 | - | 0.0 | 75.06% (429) | L-lysine aminotransferase |
| M. tuberculosis H37Rv | Rv3290c | lat | 0.0 | 100.00% (449) | L-lysine aminotransferase |
| M. leprae Br4923 | MLBr_00485 | gabT | 4e-24 | 28.47% (418) | 4-aminobutyrate aminotransferase |
| M. abscessus ATCC 19977 | MAB_3646c | - | 0.0 | 73.44% (433) | L-lysine aminotransferase |
| M. marinum M | MMAR_1243 | lat | 0.0 | 78.62% (449) | L-lysine-epsilon aminotransferase Lat |
| M. avium 104 | MAV_4262 | - | 0.0 | 77.90% (448) | L-lysine aminotransferase |
| M. smegmatis MC2 155 | MSMEG_1764 | - | 0.0 | 79.25% (429) | L-lysine aminotransferase |
| M. thermoresistible (build 8) | TH_3246 | - | 1e-26 | 27.74% (411) | PUTATIVE diaminobutyrate--2-oxoglutarate aminotransferase |
| M. ulcerans Agy99 | MUL_2644 | lat | 0.0 | 78.17% (449) | L-lysine aminotransferase |
| M. vanbaalenii PYR-1 | Mvan_1652 | - | 0.0 | 76.46% (429) | L-lysine aminotransferase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb3318c|M.bovis_AF2122/97 ---------MAAVVKSVAL-AGRPTTPDRVHEVLGRSMLVDGLDIVLDLT
Rv3290c|M.tuberculosis_H37Rv ---------MAAVVKSVAL-AGRPTTPDRVHEVLGRSMLVDGLDIVLDLT
MMAR_1243|M.marinum_M ---------MTAAVRSTVP-ACQQIEADDVQEVLGRSMLVDGFDLVLDLS
MUL_2644|M.ulcerans_Agy99 ---------MTAAVRSTVP-ACQQIEADDIQEVLGRSMLVDGFDLVLDLS
MAV_4262|M.avium_104 ---------MTAALERLAR-ARSCTGPDRVHEVLSRSILTDGFDFVLDLD
Mflv_4796|M.gilvum_PYR-GCK ---------MTAVLSRAD--RTAVIVPADVRTVLSRSILADGLDLVLDLE
Mvan_1652|M.vanbaalenii_PYR-1 ------MPLHASVRPEADA-ETDRLGPDDVHHVLARSILADGLDLVLDID
MSMEG_1764|M.smegmatis_MC2_155 ----MTALLNTPVSGSSSPGASPELAPELVRETLARSILADGFDFVLDLE
MAB_3646c|M.abscessus_ATCC_199 MTALIFPGFQAGPTHGDQATRPDPVTPDRVHDVLRRSILADGMDLVLDLE
MLBr_00485|M.leprae_Br4923 -------MTSVEQSRQLVTEIPGPVSLELAKRLNAAVPRGVGVTLPVFVT
TH_3246|M.thermoresistible__bu ---------------MSLLATTSETELPEVFSTIESEVRSYCRGWPAVME
:
Mb3318c|M.bovis_AF2122/97 RSGGSYLVDAITGRRYLDMFTFVASSALGMNPPALVDDREFHAELMQAAL
Rv3290c|M.tuberculosis_H37Rv RSGGSYLVDAITGRRYLDMFTFVASSALGMNPPALVDDREFHAELMQAAL
MMAR_1243|M.marinum_M QSAGSYLVDARTGRRYLDMFTFFASSALGMNHPGLAEDEQFRAELLEAAL
MUL_2644|M.ulcerans_Agy99 QSAGSYLVDARTGRRYLDMFTFFASSALGMNHPGLAEDEQFRAELLEAAL
MAV_4262|M.avium_104 RSRGSVLYDARDGRRYLDMFTFFASSALGMNHPALARDAEFRAELTAAAL
Mflv_4796|M.gilvum_PYR-GCK RSHGSYLVDARTGRGYLDMFTFFASSALGMNHPALADDPDFRAELAAAAV
Mvan_1652|M.vanbaalenii_PYR-1 RSHGSHLVDARTGRSYLDMFTFFASSALGMNHPGLAGDPEFRAELAAAAL
MSMEG_1764|M.smegmatis_MC2_155 ESRGSHLVDARTGERFLDMFTFFASSALGMNHSALADDPEFRAELAQAAL
MAB_3646c|M.abscessus_ATCC_199 RSHGAHLVDARDGTDYLDMFTFFASSALGMNHPMLAEDDEFRAELATAAI
MLBr_00485|M.leprae_Br4923 RAAG-GVIEDVDGNRLIDLGSGIAVTTIGNSSPRVVDAVRAQVADFTHTC
TH_3246|M.thermoresistible__bu SARGSWITDVS-GQRYLDFFAGAGALNYGHNNPKLKEP----LLKYLMSD
: * : : * :*: : . * . . : :
Mb3318c|M.bovis_AF2122/97 NKPSNSDVYSVAMARFVETFARVLGDP-ALPHLFFVEG--GALAVENALK
Rv3290c|M.tuberculosis_H37Rv NKPSNSDVYSVAMARFVETFARVLGDP-ALPHLFFVEG--GALAVENALK
MMAR_1243|M.marinum_M NKPSNADVYSVPMARFVQTFVRVLGDP-ALPHLFFVDG--GALAVENALK
MUL_2644|M.ulcerans_Agy99 NKPSNADVYSVPMARFVQTFVRVLGDP-ALPHLFFVDG--GALAVENALK
MAV_4262|M.avium_104 NKPSNSDVYSVPMARFVETFVRVLGDP-ALPHLFFVDG--GALAVENALK
Mflv_4796|M.gilvum_PYR-GCK NKPSNSDVYSVPMARFVDTFSRVLGDP-ALPHLFFVDG--GALAVENALK
Mvan_1652|M.vanbaalenii_PYR-1 NKPSNSDVYSVPMARFVDTFARVLGDP-DLLHLFFVDG--GALAVENALK
MSMEG_1764|M.smegmatis_MC2_155 NKPSNSDIYSVPMARFVDTFRRVLGDP-ALPHLFFVDG--GALAVENALK
MAB_3646c|M.abscessus_ATCC_199 NKPSNSDIYTVEMARFVDTFARVLGDP-ALPHLFFVDG--GALAVENALK
MLBr_00485|M.leprae_Br4923 FIITPYEEY----VAVTEQLNRITPGS-GEKRSVLFNS--GAEAVENSIK
TH_3246|M.thermoresistible__bu NIVHSLDMATAAKQEFLETFSELILKPRGLDYKVQFPGPTGANAVESALK
: . : : .: . . . . ** ***.::*
Mb3318c|M.bovis_AF2122/97 AAFDWKSRHNQAHGIDPALGTQVLHLRGAFHGRSGYTLSLTNTKPTITAR
Rv3290c|M.tuberculosis_H37Rv AAFDWKSRHNQAHGIDPALGTQVLHLRGAFHGRSGYTLSLTNTKPTITAR
MMAR_1243|M.marinum_M VAFDWKSRHNQALGIDAGLGTKVLHLRGAFHGRSGYTLSLTNTKPVNVAR
MUL_2644|M.ulcerans_Agy99 VAFDWKSRHNQALGIDAGLGTKVLHLRGAFHGRSGYTLSLTNTKPVNVAR
MAV_4262|M.avium_104 VAFDWKNRHNERRGVDPALGTRVLHLRGAFHGRSGYTLSLTNTKPVTVAR
Mflv_4796|M.gilvum_PYR-GCK VAFDWKSRLNEAQGRSPDLGTKVLHLRGAFHGRSGYTMSLTNTDPNKVAR
Mvan_1652|M.vanbaalenii_PYR-1 VAFDWKSRLNEARGLDPALGTKVLHLKGAFHGRSGYTLSLTNTDPNKVAR
MSMEG_1764|M.smegmatis_MC2_155 VAFDWKSRHNEAHGIDPELGTKVLHLRGAFHGRSGYTLSLTNTDPVKVAR
MAB_3646c|M.abscessus_ATCC_199 VAFDWKSRWNEAHGIDASLGTKVLHLREAFHGRSGYTMSLTNTDPNKVAR
MLBr_00485|M.leprae_Br4923 VARSYTRKP------------AVVAFDHAYHGRTNLTMALTAKSMPYKSG
TH_3246|M.thermoresistible__bu LARKVTGRE------------SIVSFTNAFHGMTLGALSVTGNSMKRAGA
* . . : :: : *:** : ::::* .. .
Mb3318c|M.bovis_AF2122/97 FPKFDWPRIDAPYMRPGLDEPAMAALEAEALRQA----RAAF-----ETR
Rv3290c|M.tuberculosis_H37Rv FPKFDWPRIDAPYMRPGLDEPAMAALEAEALRQA----RAAF-----ETR
MMAR_1243|M.marinum_M FPTFDWPRIDAPYIRP---DADMDAVEAESLAQA----RAAF-----EAH
MUL_2644|M.ulcerans_Agy99 FPTFDWPRIDAPYIRP---DADMDAVEAESLAQA----RAAF-----EAH
MAV_4262|M.avium_104 FPRFDWPRIDAPHLRCG---ADIDALEAESLRQA----RAAF-----AAH
Mflv_4796|M.gilvum_PYR-GCK FPKFDWPRIDAPSVRSGAN---IEALEAESLRQA----RAAF-----EAH
Mvan_1652|M.vanbaalenii_PYR-1 FPKFDWPRIDAPVCSAGMDNEAVDALEAESLRQA----RAAF-----EAN
MSMEG_1764|M.smegmatis_MC2_155 FPKFDWPRIDAPYVRPGLDDDAMAALEAESLRQA----RAAF-----EAH
MAB_3646c|M.abscessus_ATCC_199 FPKFDWPRIDAPYLRPG---ADIEALEASALAQA----RRAF-----AEN
MLBr_00485|M.leprae_Br4923 FGPFAPEIYRAPLSYPYRDGLLNKDLATDGKLAG----ARAINVIEKQVG
TH_3246|M.thermoresistible__bu GVPLVHS---TPMPYDNYFGGVTEDFQWFEKVLDDEGS-----------G
: :* .
Mb3318c|M.bovis_AF2122/97 PHDIACFVAEPIQGEGGDRHFRPEFFAAMRELCDEFDALLIFDEVQTGCG
Rv3290c|M.tuberculosis_H37Rv PHDIACFVAEPIQGEGGDRHFRPEFFAAMRELCDEFDALLIFDEVQTGCG
MMAR_1243|M.marinum_M PHDIACFIAEPIQGEGGDRHFRPEFFAAMRRLCDEHDALLIFDEVQTGCA
MUL_2644|M.ulcerans_Agy99 PHDIACFIAEPIQGEGGDRHFRPEFFAAMRRLCDEHDALLIFDEVQTGCA
MAV_4262|M.avium_104 PHDIACFIAEPIQGEGGDRHFRPEFFAAMRRLCDEHDALLIFDEVQTGCG
Mflv_4796|M.gilvum_PYR-GCK PHDIACFIAEPIQGEGGDRHLRPEFFAAMRELCDEFDALLIFDEVQTGCG
Mvan_1652|M.vanbaalenii_PYR-1 PHDIACFIAEPIQGEGGDRHFRPQFFAAMRQLCDEFDSLLIFDEVQTGCG
MSMEG_1764|M.smegmatis_MC2_155 PHDIACFIAEPIQGEGGDRHFRPEFFAAMRELCDEFDALLIFDEVQTGCG
MAB_3646c|M.abscessus_ATCC_199 PHDIACFIAEPIQGEGGDHHFRPEFFHAMRALCHENDALFIFDEVQTGCG
MLBr_00485|M.leprae_Br4923 ADDLAAVIIEPIQGEGGFIVPAEGFLATLLDWCRKNNVMFIADEVQTGFA
TH_3246|M.thermoresistible__bu LNRPAAVIVETVQGEGGLNVARAEWLRALAELCHTRDILLIVDDVQMGCG
. *..: *.:***** :: :: * : ::* *:** * .
Mb3318c|M.bovis_AF2122/97 LTGTAWAYQQLDVAPDIVAFGKKTQVCGVMAGRRVDEVADNVFAVPSRLN
Rv3290c|M.tuberculosis_H37Rv LTGTAWAYQQLDVAPDIVAFGKKTQVCGVMAGRRVDEVADNVFAVPSRLN
MMAR_1243|M.marinum_M MTGTPWAYQQLGVKPDVVAFGKKTQVCGVMAGGRVDEIVDNVFAVSSRLN
MUL_2644|M.ulcerans_Agy99 MTGTPWAYQQLGVKPDVVAFGKKTQVCGVMAGGRVDEIVDNVFAVSSRLN
MAV_4262|M.avium_104 LTGTAWAYQQFGVQPDVVAFGKKTQVCGIMAGRRVDEVADNVFAVSSRLN
Mflv_4796|M.gilvum_PYR-GCK ITGTAWAYQQLGVLPDVVAFGKKTQVCGVMAGTRVDEVPDNVFRTSSRIN
Mvan_1652|M.vanbaalenii_PYR-1 ITGTPWAYQQLGVRPDVVAFGKKTQVCGVMAGGRVDEVPANVFHTSSRIN
MSMEG_1764|M.smegmatis_MC2_155 ITGTAWAYQQLGVMPDVVSFGKKTQVCGVMAGRRVDEVADNVFAVSSRIN
MAB_3646c|M.abscessus_ATCC_199 LTGTAWAFQQLGVIPDVVAFGKKTQVCGVMAGGRVDEVDDNVFAVSSRIN
MLBr_00485|M.leprae_Br4923 RTGAMFACEHDGIVPDLICTAKGIADGLPLAAVTGRAEIMNAPHVSG-LG
TH_3246|M.thermoresistible__bu RTGAFFSFEEAGITPDIVTLSKSISGYGLPMALTLFRRELDVWAPGEHNG
**: :: :. .: **:: .* . :. .
Mb3318c|M.bovis_AF2122/97 STWGGNLTDMVRARRILEVIEAEGLFERAVQHGKYLRARLDELAADFPAV
Rv3290c|M.tuberculosis_H37Rv STWGGNLTDMVRARRILEVIEAEGLFERAVQHGKYLRARLDELAADFPAV
MMAR_1243|M.marinum_M STWGGNLTDMVRARRILEVIEAEGLVDNAAEQGRYLRAQFDELAKDLPAV
MUL_2644|M.ulcerans_Agy99 STWGGNLTDMVRSRRILEVIEAEGLVDNAAEQGRYLRAQFDELAKDLPAV
MAV_4262|M.avium_104 STWGGNLADMVRARRILEVIEADELFEHAARQGAYLLRGLHDLAREFPGL
Mflv_4796|M.gilvum_PYR-GCK STWGGNLVDMVRARRILEVIERDGLFDNAAQRGRHLRDGLAGLAVEFPGL
Mvan_1652|M.vanbaalenii_PYR-1 STWGGNLADMVRARRILEVIEVDGLFDRAAESGRYLRDGLDGLAADFPVL
MSMEG_1764|M.smegmatis_MC2_155 STWGGNLVDMVRSRRILEVIERDGLLDNARDMGAYLLDELRVLATRFPGV
MAB_3646c|M.abscessus_ATCC_199 STWGGNLVDMVRSRRILEVVEAQRLFANAMRMGEYLLERLQTLAVRHDAV
MLBr_00485|M.leprae_Br4923 GTFGGNPVACAAALATITTIENDGLIQRAQQIERLITDRLLRLQDADDRI
TH_3246|M.thermoresistible__bu TFRGHNPAFVTATAALKTYWQDDSFTAQTVRKGEFLRNELESIAARHEGV
* * . . : : : : .: : : : :
Mb3318c|M.bovis_AF2122/97 VLDPRGRGLMCAFSLPTTADRD------ELIRQLWQRA-VIVLPAGADT-
Rv3290c|M.tuberculosis_H37Rv VLDPRGRGLMCAFSLPTTADRD------ELIRQLWQRA-VIVLPAGADT-
MMAR_1243|M.marinum_M VLHPRGRGLMCAFDLPTTADRD------ELIQRLWQRA-VIALPAGKVS-
MUL_2644|M.ulcerans_Agy99 VLHPRGRGLMCAFDLPTTADRD------ELIQRLWQRA-VIALPAGKVS-
MAV_4262|M.avium_104 VLDVRGRGLMCAFSLPTTADRD------ELIGRLWRGG-VIVLPTGDDG-
Mflv_4796|M.gilvum_PYR-GCK VRDVRGRGLMCAFSMPSARQRD------ELLARLWECG-VIMLGCGTDS-
Mvan_1652|M.vanbaalenii_PYR-1 VRDVRGRGLMCAFSMPSAAQRD------ALIAGLWDCG-VIMLGSGPDS-
MSMEG_1764|M.smegmatis_MC2_155 VVDPRGRGLMCAFSMPTPEDRD------ALIRKLWDRH-VIMLPSGADS-
MAB_3646c|M.abscessus_ATCC_199 G-DVRGRGLMCAFSLPTAEERD------HLVQRLWADERVILLPSGENS-
MLBr_00485|M.leprae_Br4923 G-DVRGRGAMIAVELVKSGTAEPDPELTEKVATAAHATGVIILTCGMFGN
TH_3246|M.thermoresistible__bu S--ARGRGMAQGLKFEQEDLAG------AVCRAAFDRG-LLMETSGPSDE
**** ...: :: *
Mb3318c|M.bovis_AF2122/97 -VRFRPPLTVSTAEIDAAIAAVRSALPVVT-----
Rv3290c|M.tuberculosis_H37Rv -VRFRPPLTVSTAEIDAAIAAVRSALPVVT-----
MMAR_1243|M.marinum_M -VRFRPALTVGRDEIDLAIAAVRSALSAMT-----
MUL_2644|M.ulcerans_Agy99 -VRFRPALTVGRDEIDLAIAAVRSALSAMT-----
MAV_4262|M.avium_104 -VRFRPALTVSRGEIDAAIGAVRDALPTLG-----
Mflv_4796|M.gilvum_PYR-GCK -VRFRPALTVNDDEIDAALTAVAAVLRTMAVPQAG
Mvan_1652|M.vanbaalenii_PYR-1 -VRFRPALTVSTAELDAALDAVRAVLRTM------
MSMEG_1764|M.smegmatis_MC2_155 -VRFRPALPVTRAEIDACLDAVRAVLSER------
MAB_3646c|M.abscessus_ATCC_199 -VRFRPPLIVSAEEIDAAVAAVSRVLALR------
MLBr_00485|M.leprae_Br4923 IIRLLPPLTISDELLAEGLDILSRILGDF------
TH_3246|M.thermoresistible__bu VVKLLPPLTMSQEELEAGLGILADAVAATVD----
::: *.* : : : : :