For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MNQHFDVLIIGAGLSGIGTACHVTAEFPDKTIALLERRERLGGTWDLFRYPGVRSDSDMFTFGYKFRPWR DVKVLADGASIRQYIADTATEFGIDEKIHYGLKVNTAEWSSRQCRWTVAGVHEATGETRTYTCDYLISCT GYYNYDAGYLPDFPGVHRFGGRCVHPQHWPEDLDYSGKKVVVIGSGATAVTLVPAMAGSNPGSAAHVTML QRSPSYIFSLPAVDKISEVLGRFLPDRWVYEFGRRRNIAIQRKLYQACRRWPKLMRRLLLWEVRRRLGRS VDMSNFTPNYLPWDERLCAVPNGDLFKTLASGAASVVTDQIETFTEKGILCKSGREIEADIIVTATGLNI QMLGGMRLIVDGAEYQLPEKMTYKGVLLENAPNLAWIIGYTNASWTLKSDIAGAYLCRLLRHMADNGYTV ATPRDAQDCALDVGMFDQLNSGYVKRGQDIMPRQGSKHPWRVLMHYEKDAKILLEDPIDDGVLHFAAAAQ DHAAA
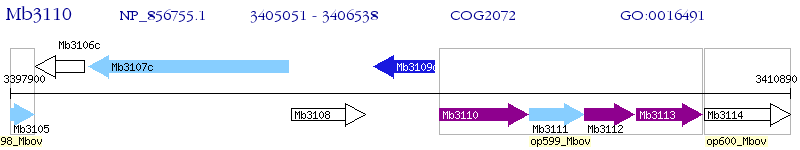
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3110 | - | - | 100% (495) | monooxygenase |
| M. bovis AF2122 / 97 | Mb3884c | ethA | e-143 | 49.80% (488) | monooxygenase ETHA |
| M. bovis AF2122 / 97 | Mb0580c | - | e-119 | 44.72% (483) | monooxygenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0962 | - | 1e-140 | 49.38% (486) | monooxygenase, flavin-binding family protein |
| M. tuberculosis H37Rv | Rv3083 | - | 0.0 | 99.80% (495) | monooxygenase |
| M. leprae Br4923 | MLBr_00065 | - | 1e-139 | 49.79% (486) | putative monooxygenase |
| M. abscessus ATCC 19977 | MAB_0985 | - | 1e-145 | 50.91% (495) | putative monooxygenase EthA |
| M. marinum M | MMAR_5404 | ethA | 1e-142 | 50.10% (491) | monooxygenase EthA |
| M. avium 104 | MAV_0175 | - | 1e-130 | 49.04% (471) | monooxygenase, flavin-binding family protein |
| M. smegmatis MC2 155 | MSMEG_6440 | - | 1e-145 | 51.23% (486) | monooxygenase, flavin-binding family protein |
| M. thermoresistible (build 8) | TH_1149 | - | 1e-136 | 51.19% (461) | MONOOXYGENASE ETHA |
| M. ulcerans Agy99 | MUL_5026 | ethA | 1e-140 | 49.49% (491) | monooxygenase EthA |
| M. vanbaalenii PYR-1 | Mvan_5683 | - | 1e-138 | 48.87% (487) | hypothetical protein Mvan_5683 |
CLUSTAL 2.0.9 multiple sequence alignment
Mflv_0962|M.gilvum_PYR-GCK -----MSDHFDVVIVGAGISGISTAWHLQQRCPDKSYVILERRENIGGTW
MSMEG_6440|M.smegmatis_MC2_155 -----MTEHFDVVIVGAGISGISTAWHLQDRCPTKSYVILERRANIGGTW
MAV_0175|M.avium_104 ---------------------MSAAWHLQNRCPTKSYAILERRADLGGTW
TH_1149|M.thermoresistible__bu ---VVMTEHLDVLIVGAGISGISAAWHLQDRCPSKSYAILERRDNLGGTW
Mvan_5683|M.vanbaalenii_PYR-1 -----MTEFVDVVIVGAGISGISAAWHLQDRCPGKSYVILERRDNLGGTW
MMAR_5404|M.marinum_M -----MTEHLDVVIVGAGISGVSAAWHLQDRCPTKSYAILEKRAAMGGTW
MUL_5026|M.ulcerans_Agy99 -----MTEHLDVVIVGAGISGVSAAWHLQDRCPTKSYAILEKRAAMGGTW
MLBr_00065|M.leprae_Br4923 MVVMAETEYLDVVIVGAGISGVSAAWHLQDRCPNKSYVILEKRAGMGGTW
MAB_0985|M.abscessus_ATCC_1997 -----MTEHVDIVIVGAGISGIGMACRVTRELPGKKFVVLEARERIGGTW
Mb3110|M.bovis_AF2122/97 -----MNQHFDVLIIGAGLSGIGTACHVTAEFPDKTIALLERRERLGGTW
Rv3083|M.tuberculosis_H37Rv -----MNQHFDVLIIGAGLSGIGTACHVTAEFPDKTIALLERRERLGGTW
:. * :: . * *. .:** * :****
Mflv_0962|M.gilvum_PYR-GCK DLFKYPGIRSDSDMFTLGFRFKPWPSARSIADGPSIWNYINEAAQENGID
MSMEG_6440|M.smegmatis_MC2_155 DLFKYPGIRSDSDMFTLGFRFKPWTSAKSIADGPSIWNYINEAAQENGID
MAV_0175|M.avium_104 DLFKYPGIRSDSDMFTLGFRFKPWRSAKSIADGASIKAYIKEAAVENRIE
TH_1149|M.thermoresistible__bu DLFKYPGIRSDSDMFTLGFRFKPWTDPKAIADGPDILAYLKETAEEFGID
Mvan_5683|M.vanbaalenii_PYR-1 DLFKYPGIRSDSDMFTLGFRFKPWTSDKAIADGPDIMAYLNETVAEYGID
MMAR_5404|M.marinum_M DLFRYPGIRSDSDMHTLGFRFRPWTERQAIADGKPILEYVKGTAAMYGID
MUL_5026|M.ulcerans_Agy99 ELFRYPGIRSDSDMHTLGFRFRPWTERQAIADGKPILEYVKGTAAMYGID
MLBr_00065|M.leprae_Br4923 DLFRYPGIRSDSDMYTLGFRFRPWTKCQAIADGKPILEYIKSTAVMHGID
MAB_0985|M.abscessus_ATCC_1997 DLFRYPGVRSDSDMFTLGYNFRPWTSTKGIADGTSIREYVTETAREFGVT
Mb3110|M.bovis_AF2122/97 DLFRYPGVRSDSDMFTFGYKFRPWRDVKVLADGASIRQYIADTATEFGID
Rv3083|M.tuberculosis_H37Rv DLFRYPGVRSDSDMFTFGYKFRPWRDVKVLADGASIRQYIADTATEFGVD
:**:***:******.*:*:.*:** . : :*** * *: :. :
Mflv_0962|M.gilvum_PYR-GCK KHIRVKHQVLAANWSDAENRWELT--VDHDGEQKQITCSFLSVCSGYYNY
MSMEG_6440|M.smegmatis_MC2_155 KHIRTNHRVLGADWSDAENRWTIT--VEADGEQKQITASFLSVCSGYYNY
MAV_0175|M.avium_104 PHIRYRHRVVAADWSDADNRWTVT--VEHDGQRSEITCSFLFACTGYYNY
TH_1149|M.thermoresistible__bu EHIRYRHRVVSANWSDPDNKWILQ--VERDGEQVEMTCSFLFACSGYYNY
Mvan_5683|M.vanbaalenii_PYR-1 KHIRYGQNVKSADWSDADNRWTVR--VDRAGEEVEISARYLMACSGYYNY
MMAR_5404|M.marinum_M KHIRLNQKVISADWSNAQNRWTLQ--IESNGATSAVTCSFLFLCSGYYNY
MUL_5026|M.ulcerans_Agy99 KHIRLNQKVISADWSNAQNRWTLQ--IESNGAMSAVTCSFLFLCSGYYNY
MLBr_00065|M.leprae_Br4923 KYIRLNHKVTGADWSSIENRWTVQ--VENNGTPRMISCSFLFLCSGYYNY
MAB_0985|M.abscessus_ATCC_1997 EKVRFGHKVVGAEWSSERGLWTLQ--VERAGETVEFTTQFLLGCTGYYRY
Mb3110|M.bovis_AF2122/97 EKIHYGLKVNTAEWSSRQCRWTVAGVHEATGETRTYTCDYLISCTGYYNY
Rv3083|M.tuberculosis_H37Rv EKIHYGLKVNTAEWSSRQCRWTVAGVHEATGETRTYTCDYLISCTGYYNY
:: .* *:**. * : : * : :* *:***.*
Mflv_0962|M.gilvum_PYR-GCK DQGYSPEFPGADDFAGQIIHPQHWPEDLDYTGKRIVVIGSGATAVTLIPS
MSMEG_6440|M.smegmatis_MC2_155 DQGYSPEFPGADDFAGQIIHPQHWPEDLDYAGKKIVVIGSGATAVTLIPS
MAV_0175|M.avium_104 DEGYSPTFPGAEDFGGTIVHPQHWPEDLDYAGKRIVVIGSGATAITLIPA
TH_1149|M.thermoresistible__bu DEGYSPEFPGADDFEGTIVHPQHWPEDLDYAGKRIVVIGSGATAVTLIPA
Mvan_5683|M.vanbaalenii_PYR-1 DEGYSPDFPGSEDFEGTLIHPQHWPEDLDYAGKRIVVIGSGATAVTLIPA
MMAR_5404|M.marinum_M DQGYTPNFAGAQDFAGSIIHPQHWPEDLDYDGKNVVVIGSGATAVTLVPA
MUL_5026|M.ulcerans_Agy99 DQGYTPNFAGAQDFAGSIIHPQHWPEDLDYDGKNVVVIGSGASAVTLVPA
MLBr_00065|M.leprae_Br4923 EQGYAPTFLGSEDFTGPIIHPQHWPEDLDYAAKNIVVIGSGATAVTLVPA
MAB_0985|M.abscessus_ATCC_1997 DEGFTPKFEGIEDFAGQVVHPQHWPEDLDYSGKRVVVIGSGATAMTLVPA
Mb3110|M.bovis_AF2122/97 DAGYLPDFPGVHRFGGRCVHPQHWPEDLDYSGKKVVVIGSGATAVTLVPA
Rv3083|M.tuberculosis_H37Rv DAGYLPDFPGVHRFGGRCVHPQHWPEDLDYSGKKVVVIGSGATAVTLVPA
: *: * * * . * * :*********** .*.:*******:*:**:*:
Mflv_0962|M.gilvum_PYR-GCK LVAGG---AGHVTMLQRSPTYIGSLPLVDPIAEKANKYLPGSLAHFVNRW
MSMEG_6440|M.smegmatis_MC2_155 LVNGG---AAHVTMLQRSPTYIGSLPLVDPVAEKTNKYLPKNLAHFVNRW
MAV_0175|M.avium_104 LVNSG---AGHVTMLQRSPTYIGSLPGVDPFAERANRLLPDRLAHMANRW
TH_1149|M.thermoresistible__bu LAETG---AGHVTMLQRSPTYIGSLPLVDPIAVKVNKYLPPKMASVVNRW
Mvan_5683|M.vanbaalenii_PYR-1 LANSG---AGHVTMLQRSPTFIAALPDVDPFTVATNKRLPAKVAYVLNRW
MMAR_5404|M.marinum_M LANSG---AKHVTMLQRSPTYIVSQPERDSIAERLNRWLPENMAYAMVRW
MUL_5026|M.ulcerans_Agy99 LANSG---AKHVTMLQRSPTYIVSQPERDSIAERLNRWLPENMAYAMVRW
MLBr_00065|M.leprae_Br4923 LANSG---AKHVTMLQRSPTYIVSQPAKDKIAARLNRWLPDKYAYTAVRW
MAB_0985|M.abscessus_ATCC_1997 MAGT----AGHVTMLQRSPTYVVSLPSTDALAVALQGKLPASLAYPIVKW
Mb3110|M.bovis_AF2122/97 MAGSNPGSAAHVTMLQRSPSYIFSLPAVDKISEVLGRFLPDRWVYEFGRR
Rv3083|M.tuberculosis_H37Rv MAGSNPGSAAHVTMLQRSPSYIFSLPAVDKISEVLGRFLPDRWVYEFGRR
:. * *********::: : * * .: ** . :
Mflv_0962|M.gilvum_PYR-GCK KAISFSTLQYQLSQRFPKYMRKTLMTMAQRRLPEGYDVEKHFGPRYNPWD
MSMEG_6440|M.smegmatis_MC2_155 KAIAFSTAQYQLARKFPNYMRKTLMTMAQRRLPEGFDVQKHFGPRYNPWD
MAV_0175|M.avium_104 KAIAFSTFQYQLSRKAPAYMRKTLMTMAKRRLPEGYDVEKHFGPRYNVWD
TH_1149|M.thermoresistible__bu KAIIFSTFQYQLSRKFPNYMRRTLMRMAERRLPPGYDVEKHFGPRYNPWD
Mvan_5683|M.vanbaalenii_PYR-1 KSILQQSVQYQVARKFPDFFRKQLLTMAKHRLPEGYDVNKHFNPRYNPWD
MMAR_5404|M.marinum_M KNVLRQAAVYGACRKWPRRMRKNFMGLAQRQLPEGYDVRKHFGPHYNPWD
MUL_5026|M.ulcerans_Agy99 KNVLRQAAVYGACRKWPRRMRKNFMGLAQRQLPEGYDVRKHFGPHYNPWD
MLBr_00065|M.leprae_Br4923 KNILRQSALYGACQKWPQRMRNILLGHVARRLPKGYDVQKHFGPHYNPWE
MAB_0985|M.abscessus_ATCC_1997 KNQMVSFISYQTSRRDPERMKGILRALLKRQLPN-FDLDKHFTPKYNPWD
Mb3110|M.bovis_AF2122/97 RNIAIQRKLYQACRRWPKLMRRLLLWEVRRRLGRSVDMSN-FTPNYLPWD
Rv3083|M.tuberculosis_H37Rv RNIAIQRKLYQACRRWPKLMRRLLLWEVRRRLGRSVDMSN-FTPNYLPWD
: . * .:: * :: : ::* *: : * *.* *:
Mflv_0962|M.gilvum_PYR-GCK ERLCLAPNGDLFKTIRAGKADVVTDTIDRFTETGIRLNSGEELAADIIIT
MSMEG_6440|M.smegmatis_MC2_155 ERLCLAPNGDLFKTIRAGKADVVTDTIAKFTETGIKLTSGEELTADIIIT
MAV_0175|M.avium_104 ERLCLAPDGDFFRTIRHGKADVVTDTIDRFTTTGIRLNSGEELPADIIVT
TH_1149|M.thermoresistible__bu ERLCLAPNGDLFRAIRHGKADVVTDTIDTFTKTGIRLTSGTELEADIIIT
Mvan_5683|M.vanbaalenii_PYR-1 ERVCLAPNGDLFKTIRAGKADVVTDTIERFTKTGIQLASGRHLDADIIVT
MMAR_5404|M.marinum_M QRLCLVPNGDLFRAIRHGQVDVVTDTIDRFTPTGIRLNSGQQLPADIIVT
MUL_5026|M.ulcerans_Agy99 QRLCLVPNGDLFRAIRHGQVDVVTDTIDRFTPTGIRLNSGQQLPADIIVT
MLBr_00065|M.leprae_Br4923 QRLCLVPDGDLFRAIRKGTVDMVTDAIDRFTSTGIRLKSGNELRADIIVT
MAB_0985|M.abscessus_ATCC_1997 QRLCVVPDSDLFRALRKGTASIVTDRITRFTPKGILLESGEELEADIVVT
Mb3110|M.bovis_AF2122/97 ERLCAVPNGDLFKTLASGAASVVTDQIETFTEKGILCKSGREIEADIIVT
Rv3083|M.tuberculosis_H37Rv ERLCAVPNGDLFKTLASGAASVVTDQIETFTEKGILCKSGREIEADIIVT
:*:* .*:.*:*::: * ..:*** * ** .** ** .: ***::*
Mflv_0962|M.gilvum_PYR-GCK ATGLNMQLFGGATATRNGEPIDLTKTMTYKGLMLSGVPNMAITFGYTNAS
MSMEG_6440|M.smegmatis_MC2_155 ATGLNMQLFGGASLTRNGQEVDLTETMTYKGLMLSGVPNMAITFGYTNAS
MAV_0175|M.avium_104 ATGLNMQLLGGVTPTRNGDPVDLTSLMTYKGLMFSGMPNFAITFGYTNAS
TH_1149|M.thermoresistible__bu ATGLNLQLFGGAEIKRNGQPVELTETMAYKGMMLSGMPNMAFTIGYTNAS
Mvan_5683|M.vanbaalenii_PYR-1 ATGLNLQLFGGATISRNGEAIELNDTMAYKGMLLTDMPNLAFTIGYTNAS
MMAR_5404|M.marinum_M ATGLNLQLFGGAAVTVDGESVDLTKAMAYKGMMLSGLPNMAYTVGYTNAS
MUL_5026|M.ulcerans_Agy99 ATGLNLQLFGGAAVTVDGESVDLTKAMAYKGMMLSGLPNMAYTVGYTNAS
MLBr_00065|M.leprae_Br4923 ATGLNLQLFGGAVATVDGQPVDLAQTMSYKGMMLSGLPNMIYTVGYTNAS
MAB_0985|M.abscessus_ATCC_1997 ATGLNVQLAGGLRPIVDGVQLNPADSVSYKGLMLTGLPNFIFTFGYTNAS
Mb3110|M.bovis_AF2122/97 ATGLNIQMLGGMRLIVDGAEYQLPEKMTYKGVLLENAPNLAWIIGYTNAS
Rv3083|M.tuberculosis_H37Rv ATGLNIQMLGGMRLIVDGAEYQLPEKMTYKGVLLENAPNLAWIIGYTNAS
*****:*: ** :* : . ::***::: . **: .******
Mflv_0962|M.gilvum_PYR-GCK WTLKADLVSEFICRLLNYMDDNGFDRVEAQHPGD-SVEEKPFMDFSPGYF
MSMEG_6440|M.smegmatis_MC2_155 WTLKADLVSEFICRVLNYMDDNGFDRVEPQHPGD-AVDALPFMDFNPGYF
MAV_0175|M.avium_104 WTLKADLVSEFVCRLLNYMDAKGFDFVEPQHPGE-DVDALPFMDFTPGYF
TH_1149|M.thermoresistible__bu WTLKADLVSEFVCRVLNHMDEHGYDVVEPQHPGD-SVEERPLLDFTPGYV
Mvan_5683|M.vanbaalenii_PYR-1 WTLKADLVSEFFCRIINYMDANSYDRVEPRHPGS-SVDERPLMDFTPGYV
MMAR_5404|M.marinum_M WTLKADLVSEFVCRLLNYMDANDYDSVVVERPGS-DVQECPFMEFTPGYV
MUL_5026|M.ulcerans_Agy99 WTLKADLVSEFVCRLLNYMDANDYESVVVERPGS-DVQECPFMEFTPGYV
MLBr_00065|M.leprae_Br4923 WTLKADLVSEFFCRLLNYMNANDFDTVAVEYPGS-DVEERPFMDFTPGYV
MAB_0985|M.abscessus_ATCC_1997 WTLRADLVSQYTCRLLKYMDKEGQTVVWPVEPDT-AERLPFLEDLVSGYV
Mb3110|M.bovis_AF2122/97 WTLKSDIAGAYLCRLLRHMADNGYTVATPRDAQDCALDVGMFDQLNSGYV
Rv3083|M.tuberculosis_H37Rv WTLKSDIAGAYLCRLLRHMADNGYTVATPRDAQDCALDVGMFDQLNSGYV
***::*:.. : **::.:* .. . . : :: .**.
Mflv_0962|M.gilvum_PYR-GCK RRAMDTLPKSGSEAPWR--LKQNYFIDLRTIRYGKVDEESLRFTKHGAPV
MSMEG_6440|M.smegmatis_MC2_155 RRAMDSLPKSGSRAPWR--LKQNYFFDLRMIRYDKVDEESLHFTKHRAAV
MAV_0175|M.avium_104 RRSMHLLPKSGSRAPWR--LKQNYFFDMRTIRRGRVDDEGLKFAKKRAPV
TH_1149|M.thermoresistible__bu LRALDRLPPPXXXRMWWTRCRRSRFAAGPPAWRSTTRRRSSRPRPGRWGS
Mvan_5683|M.vanbaalenii_PYR-1 LRALDYLPKAGSRAPWR--LKQNYLMDLQLIRRGKVDDEALAFSRHPAQV
MMAR_5404|M.marinum_M LRVIDELPKQGSRKPWR--LNQNYLRDIHLIRRGKIDDEGLRFAKKPVAV
MUL_5026|M.ulcerans_Agy99 LRVIDELPKQGSRKPWR--LNQDYLRDIHLIRRGKIDDEGLRFAKKPVAV
MLBr_00065|M.leprae_Br4923 LRSLDALPKQGSRTPWR--LNQNYLRDIRLIRRGKVADEGLLFAKKTTPV
MAB_0985|M.abscessus_ATCC_1997 TRAENAMPHQVPLAPWR--SYQNYFRELPVLKYGKVADKGVRFSSGAAGS
Mb3110|M.bovis_AF2122/97 KRGQDIMPRQGSKHPWR--VLMHYEKDAKILLEDPIDDGVLHFAAAAQDH
Rv3083|M.tuberculosis_H37Rv KRGQDIMPRQGSKHPWR--VLMHYEKDAKILLEDPIDDGVLHFAAAAQDH
* . :* * .
Mflv_0962|M.gilvum_PYR-GCK ALSS--------------
MSMEG_6440|M.smegmatis_MC2_155 SASSS-------------
MAV_0175|M.avium_104 AV----------------
TH_1149|M.thermoresistible__bu TGCG--------------
Mvan_5683|M.vanbaalenii_PYR-1 TASA--------------
MMAR_5404|M.marinum_M PV----------------
MUL_5026|M.ulcerans_Agy99 PV----------------
MLBr_00065|M.leprae_Br4923 TV----------------
MAB_0985|M.abscessus_ATCC_1997 ATSGRGNASHEAPVAIGR
Mb3110|M.bovis_AF2122/97 AAA---------------
Rv3083|M.tuberculosis_H37Rv AAA---------------
.