For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSQHDPVSAAWRAHRAYLVDLAFRMVGDIGVAEDMVQEAFSRLLRAPVGDIDDERGWLIVVTSRLCLDHI KSASTRRERPQDIAAWHDGDASVSSVDPADRVTLDDEVRLALLIMLERLGPAERVVFVLHEIFGLPYQQI ATTIGSQASTCRQLAHRARRKINESRIAASVEPAQHRVVTRAFIEACSNGDLDTLLEVLDPGVAGEIDAR KGVVVVGADRVGPTILRHWSHPATVLVAQPVCGQPAVLAFVNRALAGVLALSIEAGKITKIHVLVQPSTL DPLRAELGGG
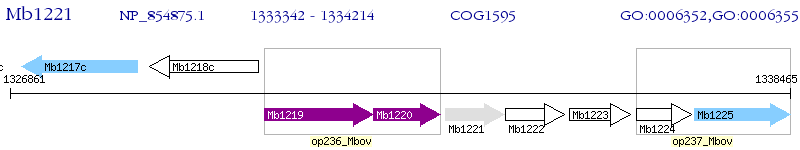
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1221 | sigI | - | 100% (290) | RNA polymerase sigma factor SigI |
| M. bovis AF2122 / 97 | Mb3361c | sigJ | 2e-28 | 36.04% (222) | RNA polymerase sigma factor SigJ |
| M. bovis AF2122 / 97 | Mb0188c | sigG | 4e-11 | 26.82% (220) | RNA polymerase factor sigma-70 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_1927 | - | 1e-30 | 29.93% (274) | ECF subfamily RNA polymerase sigma-24 factor |
| M. tuberculosis H37Rv | Rv1189 | sigI | 1e-164 | 100.00% (290) | RNA polymerase sigma factor SigI |
| M. leprae Br4923 | MLBr_01076 | sigE | 2e-12 | 30.82% (159) | RNA polymerase sigma factor SigE |
| M. abscessus ATCC 19977 | MAB_2999c | sigI | 2e-58 | 46.34% (287) | RNA polymerase sigma factor SigI |
| M. marinum M | MMAR_4487 | - | 5e-33 | 31.64% (275) | alternative RNA polymerase sigma factor |
| M. avium 104 | MAV_0164 | sigI | 5e-63 | 49.65% (284) | RNA polymerase sigma factor SigI |
| M. smegmatis MC2 155 | MSMEG_5444 | - | 1e-29 | 31.18% (279) | RNA polymerase sigma-70 factor protein |
| M. thermoresistible (build 8) | TH_1142 | - | 2e-30 | 30.58% (278) | RNA polymerase sigma factor, sigma-70 family protein |
| M. ulcerans Agy99 | MUL_3630 | - | 9e-28 | 31.47% (286) | alternative RNA polymerase sigma factor SigJ- like |
| M. vanbaalenii PYR-1 | Mvan_4806 | - | 6e-30 | 29.93% (274) | ECF subfamily RNA polymerase sigma-24 factor |
CLUSTAL 2.0.9 multiple sequence alignment
Mb1221|M.bovis_AF2122/97 -------MSQHDPVSAAWRAHRAYLVDLAFRMVGDIGVAEDMVQEAFSRL
Rv1189|M.tuberculosis_H37Rv -------MSQHDPVSAAWRAHRAYLVDLAFRMVGDIGVAEDMVQEAFSRL
MAB_2999c|M.abscessus_ATCC_199 ---MSSSSTHDALVTAAWREHYPYLVNLAYQMLGDIGDAEDAVQEAFVRL
MAV_0164|M.avium_104 ---MNRSQSADDQVAEAWRRHRPYLVNLAYQMLGDIGDAEDVAQEAFLRL
MUL_3630|M.ulcerans_Agy99 --------MTDPALLARFQAARPRLGALAYRMLGSLADAEDVVQEAWLRL
Mflv_1927|M.gilvum_PYR-GCK -----MTAAPGDEHAERFTQLRPLLFTIAYEILGSATESDDVLQESYLRW
Mvan_4806|M.vanbaalenii_PYR-1 ----------MSEHADRFTHLRPLLFTIAYEILGSATESDDVLQESYLRW
MSMEG_5444|M.smegmatis_MC2_155 --MNNPSAGNSTEHAERFTALRPLLFTIAYEILGTATESDDVLQESYLRW
TH_1142|M.thermoresistible__bu MTGSGAPSARGDEHAERFTHLRPLLFTIAYEILGSATEADDVLQESYLRW
MMAR_4487|M.marinum_M ----MTPPSHSDEHAERFTLLRPLLFTIAYEILGSATESDDVLQESYLRW
MLBr_01076|M.leprae_Br4923 -MGDGIGMEREGRWTGNTQCPLRVVPGDESPTLDGRASPEDLIITNLLSP
: :. .:*
Mb1221|M.bovis_AF2122/97 LRAPVG-----DIDDERGWLIVVTSRLCLDHIKSASTRRERPQ--DIAAW
Rv1189|M.tuberculosis_H37Rv LRAPVG-----DIDDERGWLIVVTSRLCLDHIKSASTRRERPQ--DIAAW
MAB_2999c|M.abscessus_ATCC_199 ARAEET-----EIDEPRAWLSVVVGRLCLDQLRSARMRRERTGVASVVES
MAV_0164|M.avium_104 SAAGD-------IDDVRGWLTVVASRLCLDQLRSARARHERPG--DVGEQ
MUL_3630|M.ulcerans_Agy99 NATTDRPEGAQELVNLDAWLTTVVARLCLNLLRDRRGRGREELVGQLPDP
Mflv_1927|M.gilvum_PYR-GCK AEVDLA-----TVTDTKAYLARLVTRQALNTLRAQSRRREDYVGPWLPEP
Mvan_4806|M.vanbaalenii_PYR-1 AEVDLA-----AVTDTKAYLARLVTRQALNALRAQSRRREDYVGPWLPEP
MSMEG_5444|M.smegmatis_MC2_155 AEVDLA-----TVTDTKAYLAKLVTRQSLNALRARSRLREEYVGPWLPEP
TH_1142|M.thermoresistible__bu AEVDLA-----AVRDTKSYLAKLVSRQALNALRAGARRREDYVGPWLPEP
MMAR_4487|M.marinum_M AAVDLS-----AVRDTKSYLAQLVTRQSLNALRAGARRREEYVGPWLPEP
MLBr_01076|M.leprae_Br4923 TIMSHP-----PPSRDDDWVEPFDALQGTAVFDATGDKATMPSWEELVRQ
:: . : :
Mb1221|M.bovis_AF2122/97 HDGDASVSSVDPADRVTLDDEVRLALLIMLERLGPAERVVFVLHEIFGLP
Rv1189|M.tuberculosis_H37Rv HDGDASVSSVDPADRVTLDDEVRLALLIMLERLGPAERVVFVLHEIFGLP
MAB_2999c|M.abscessus_ATCC_199 AAPLHVPAVTDPADRVTLDDEVRGALLEVLRSLSPGERVAFVLHDVFAVP
MAV_0164|M.avium_104 PAPPRF----DPADRITLDDEVRTALLEVLRRLSPGERVSFVLHDVFGVP
MUL_3630|M.ulcerans_Agy99 --IVDGVAAFDPEHHAMLADAVGLALFVVLDTLAPAERLAFVLHDVFAVP
Mflv_1927|M.gilvum_PYR-GCK LLTEA-----DPSADVVLAESVSMAMLVVLETLSPDERAVFVLREVFGFG
Mvan_4806|M.vanbaalenii_PYR-1 LLTEA-----DPSADVVLAESVSMAMLVVLETLSPDERAVFVLREVFGFG
MSMEG_5444|M.smegmatis_MC2_155 LLVAE-----DASSDVVLAESVSMAMLVVLETLTPDERAVFVLHDVFGFS
TH_1142|M.thermoresistible__bu LLLEER----DASADVVLAESVSMAMMVVLESLTPDERAVFVLREVFGFE
MMAR_4487|M.marinum_M LLLDES----DPSADLVLAESVSMAMLVLLETLTPDERAVFVLREVFGFD
MLBr_01076|M.leprae_Br4923 HAARVYWLAYRLSGNQHDAEDLTQETFIRVFRSVQNYQPGTFEGWLHRIT
: : : : : . :. .
Mb1221|M.bovis_AF2122/97 YQQIATTIGSQASTCRQLAHRARRKINESR-IAASVEPAQHRVVTRAFIE
Rv1189|M.tuberculosis_H37Rv YQQIATTIGSQASTCRQLAHRARRKINESR-IAASVEPAQHRVVTRAFIE
MAB_2999c|M.abscessus_ATCC_199 FEEIASTVGRPVGTCRQLARRARTRFQQSAARSTDVTGAEHQQLIEKFVT
MAV_0164|M.avium_104 FEAIAQTVGRPVGTCRQLARRARAKFVAAQPKLNTVGAAEHQRVTEKFIT
MUL_3630|M.ulcerans_Agy99 FDQIAPILERTPEATRKLASRARRRIERAE-PFPDRDLCAQREVVDAFFA
Mflv_1927|M.gilvum_PYR-GCK HDEIAATLGKSTAAVRQIAHRAREHVQSRRKRFEPVDPKTSGEITMTFFA
Mvan_4806|M.vanbaalenii_PYR-1 HDEIAATLGKSTEAVRQTAHRARAHVQSRRKRFEPVDPKVSTEITTRFFT
MSMEG_5444|M.smegmatis_MC2_155 HDEIASTIGKSAAAVRQMAHRARNHVQARRKRFEPVDPQVSMEITQRFFT
TH_1142|M.thermoresistible__bu YGEIATTVGKSVTAVRQIAHRARSHVHARRKRFEPVDPEVSMEITERFLT
MMAR_4487|M.marinum_M YDEIADAVGKPAPTVRQVAHRARAHIQARRKKYGDVDPQRNAQLTAQFLQ
MLBr_01076|M.leprae_Br4923 TNLFLDMVRR----------RARIRMDALPDDYDRVP-ADEPNPEQIYHD
: : *** :. :
Mb1221|M.bovis_AF2122/97 ACSNGDLDTLLEVLDPGVAG-----EIDARKGVVVVGADRVG---PTILR
Rv1189|M.tuberculosis_H37Rv ACSNGDLDTLLEVLDPGVAG-----EIDARKGVVVVGADRVG---PTILR
MAB_2999c|M.abscessus_ATCC_199 ACAQGDLTGLMAVLDPGVWGVGTILGDPAPPPQVNRGRTDVA---TNLLV
MAV_0164|M.avium_104 ACANGDLAGLAAVLDPTVWGVGTVLADPAPPPQVNHGPDAVA---TNLLR
MUL_3630|M.ulcerans_Agy99 AGRSGNFERLVSVLHPDVVLRG---DFGPNTPRFRAQGAATV---AKLAR
Mflv_1927|M.gilvum_PYR-GCK AAATGDLDALMAMLAPDVVWTADSDGKVSAARRPVEGADKVARLILGFVR
Mvan_4806|M.vanbaalenii_PYR-1 AAATGDLDGLMAMLAPDVVWTADSDGKVSAARRPVAGADRVARLIMGFVR
MSMEG_5444|M.smegmatis_MC2_155 AASTGDIDALMELLAPDVVWTADSAGKRSAARRPVSGADAVARLVSGLVR
TH_1142|M.thermoresistible__bu AASTGDMDGLLAMLAPDATWTADGGGKATAARRPIVGAEKVAAVILGIFR
MMAR_4487|M.marinum_M TAASGDVEALMAMLAPDATWLADSGGKVSAARKPVVGAERVARAITGLIR
MLBr_01076|M.leprae_Br4923 SRLGPDLQAALDSLPPEFRAAVVLCDIEGLSYEEIG---------ATLGV
: :. * * :
Mb1221|M.bovis_AF2122/97 HWSH-PATVLVAQPVCGQPAVLAFVNRALAGVLALSIEAGKITKIHVLVQ
Rv1189|M.tuberculosis_H37Rv HWSH-PATVLVAQPVCGQPAVLAFVNRALAGVLALSIEAGKITKIHVLVQ
MAB_2999c|M.abscessus_ATCC_199 YLG--PGATLVGGP-FGQPVLLAFTQRRLFAVVTLTVREGLVLKIEATAD
MAV_0164|M.avium_104 YLW--PDVTLVGGP-AGAPVLLAFSRRRLFAVIVLSIRDARVVKIEAIAD
MUL_3630|M.ulcerans_Agy99 SYAG-PERTVLNTLVNGAAGAVISISEQASAVMRFLVRNGRIAAIDVLAD
Mflv_1927|M.gilvum_PYR-GCK LAG--GEARVEPAFYNSAPALVLYTGDVLEGVVTFEVADGRITNFYAMRN
Mvan_4806|M.vanbaalenii_PYR-1 QAG--ADGRVEPAVYNSAPALVLYLGDNLEGVVTFEVVDGRITNFYAMRN
MSMEG_5444|M.smegmatis_MC2_155 LGG--EGARVEPAMYNNAPALKLYLGDSFEGVITIEIVDGRITHFYAMRN
TH_1142|M.thermoresistible__bu AGQRLPDVRIETAIYNCAPAVVVYSRDNLEGVFTVEVVDGRITHFYAVRN
MMAR_4487|M.marinum_M KAA--TELQVELVTCNSAPAVLFYLGGQLEGVITLEIVGDKITNFYVMRN
MLBr_01076|M.leprae_Br4923 KLG-----TVRSRIHRGRQALRDY-------LLTAHPRDGATTDATHVKS
: :. .
Mb1221|M.bovis_AF2122/97 PSTLDPLRAELGGG---
Rv1189|M.tuberculosis_H37Rv PSTLDPLRAELGGG---
MAB_2999c|M.abscessus_ATCC_199 P-------AARGR----
MAV_0164|M.avium_104 P-------AARSAG---
MUL_3630|M.ulcerans_Agy99 PQRIATLDISGLSGPGG
Mflv_1927|M.gilvum_PYR-GCK PEKLTGVMVARTISR--
Mvan_4806|M.vanbaalenii_PYR-1 PEKLAGVKVPRQISR--
MSMEG_5444|M.smegmatis_MC2_155 PDKLAGVDIPRVISR--
TH_1142|M.thermoresistible__bu PEKLAAVDTPRQISR--
MMAR_4487|M.marinum_M PDKLTAVARAHEISRG-
MLBr_01076|M.leprae_Br4923 A----------------
.