For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MVVVGTDAHKYSHTFVATDEVGRQLGEKTVKATTAGHATAIMWAREQFGLELIWGIEDCRNMSARLERDL LAAGQQVVRVPTKLMAQTRKSARSRGKSDPIDALAVARAVLRETDLPLATHDETSRELKLLTDRRDVLVA QRTSAINRLRWLVHELDPERAPAARSLDAAKHQQALRTWLDTQPGLVAELARAELTDIIRLTGEINTLAQ RISARVHQVAPALLEIPGCAELTAAKIVGEAAGVTRFKSEAAFACHAAVAPIPVWSGNTAGQMRLSRSGN RQLNAALHRIALTQIRMTDSRGQAYYQRLQDAGKTKRAALRCLKRRLARTVFQALRTVHQPSSEHTQPAA ACHRSYCSRSCLSG
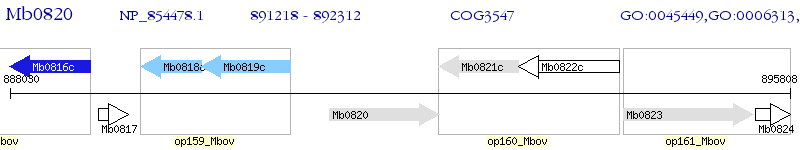
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0820 | - | - | 100% (364) | IS1547 transposase |
| M. bovis AF2122 / 97 | Mb3360 | - | 0.0 | 100.00% (358) | transposase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_0450 | - | 1e-126 | 68.25% (337) | transposase IS116/IS110/IS902 family protein |
| M. tuberculosis H37Rv | Rv0797 | - | 0.0 | 99.73% (364) | IS1547 transposase |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_3792 | - | 2e-30 | 31.91% (351) | transposase |
| M. avium 104 | MAV_1059 | - | 1e-140 | 71.79% (351) | putative transposase |
| M. smegmatis MC2 155 | MSMEG_6023 | - | 4e-31 | 33.43% (341) | transposase |
| M. thermoresistible (build 8) | TH_3017 | - | 1e-21 | 28.86% (350) | PUTATIVE transposase |
| M. ulcerans Agy99 | - | - | - | - | - |
| M. vanbaalenii PYR-1 | Mvan_0416 | - | 3e-18 | 25.87% (402) | transposase IS116/IS110/IS902 family protein |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_3792|M.marinum_M MIVAEAFGYVVGIDTHARTHTYCILNARTGAIVVGATFPNTSPAHRRVIA
MSMEG_6023|M.smegmatis_MC2_155 MTPSQIV--VIGADTHLDTIHLAAL-SETGKPLGDAEFPTRPSGYYVAVK
Mb0820|M.bovis_AF2122/97 -------MVVVGTDAHKYSHTFVAT-DEVGRQLGEKTVKATTAGHATAIM
Rv0797|M.tuberculosis_H37Rv -------MVVVGTDAHKYSHTFVAT-DEVGRQLGEKTVKATTAGHATAIM
MAV_1059|M.avium_104 -------MVVVGADVHKHTHTFVAV-DEVGRKLGEKTVKALTSGHAEAVM
Mflv_0450|M.gilvum_PYR-GCK -------MVIVGTDVHKRTHTFVAV-DEVGRKLGEKVVPATTAGHRTAVA
TH_3017|M.thermoresistible__bu -----------------------VVIDAQGKRLLSQRVANDEATLLKLIS
Mvan_0416|M.vanbaalenii_PYR-1 ----MTTGQVWAGVDVGKEHHWVCAVDDSGKVVLSRRLVNDEQPIRELVA
* : . :
MMAR_3792|M.marinum_M WIQRHCPNTKVLAAVEGTSSYGAGVTAALVDAGIDVTEV-----RPTGRH
MSMEG_6023|M.smegmatis_MC2_155 WAQSFG--TVTLAGVEGTNSYGAGLTRALQDGGIDVVEVN----RPDRAA
Mb0820|M.bovis_AF2122/97 WAREQFG-LELIWGIEDCRNMSARLERDLLAAGQQVVRVPTKLMAQTRKS
Rv0797|M.tuberculosis_H37Rv WAREQFG-LELIWGIEDCRNMSARLERDLLAAGQQVVRVPTKLMAQTRKS
MAV_1059|M.avium_104 WARERFG-AQVVWAIEDCRHLSARLERDLLTAGQQVVRVPPKLMAQARAS
Mflv_0450|M.gilvum_PYR-GCK WAREQFG-DELLWGIEDCRNLSARLEIDLLGMAQSAVRVAPKLMSQSRAC
TH_3017|M.thermoresistible__bu TVATMADGGEVTWAIDLNAGGAALLITLLIAADQRLLYIPGRTVHHASGS
Mvan_0416|M.vanbaalenii_PYR-1 EIDELAER--VSWTVDLTTVYAALVLTVLADAGKTVRYLAGRAVWQASAT
:: .* : * :
MMAR_3792|M.marinum_M RAR-AGKSDVIDAEAAARSVLGCEQQRLPIPRESGEREDLRILLAARRIL
MSMEG_6023|M.smegmatis_MC2_155 RRR-RGKSDPLDAYAAARTALSG--HGLAAPKDE-RTLALSALLTARRGA
Mb0820|M.bovis_AF2122/97 ARS-RGKSDPIDALAVARAVLRE--TDLPLATHDETSRELKLLTDRRDVL
Rv0797|M.tuberculosis_H37Rv ARS-RGKSDPIDALAVARAVMRE--TDLPLATHDETSRELKLLTDRRDVL
MAV_1059|M.avium_104 ART-RGKSDPIDALAVARGFLRE--PDLPVASHDKVSRELKLLVDRREVL
Mflv_0450|M.gilvum_PYR-GCK ART-RGKSDPIDALAVAHAVLRH--PDLPVAAHDEVSRELKLLTDRREDL
TH_3017|M.thermoresistible__bu YRG-EGKTDAKDAAVIADQARMR-RDLQPLRPGDDITVELRILTSRRTDL
Mvan_0416|M.vanbaalenii_PYR-1 YRGGEAKTDAKDARVIADQARMRGQDLPVLHPDDDLISELRMLTGHRADL
.*:* ** . * . * * *
MMAR_3792|M.marinum_M DQQRTANRNALTALLRTI--DLGIDARKPLTNDQVRVIAAWRG-------
MSMEG_6023|M.smegmatis_MC2_155 VKAHTAATNQIQSLLVTAPSELRECYRRYTTRGLVKELARCRP-------
Mb0820|M.bovis_AF2122/97 VAQRTSAINRLRWLVHELDPERAPAARSLDAAKHQQALRTWLD-------
Rv0797|M.tuberculosis_H37Rv VAQRTSAINRLRWLVHELDPERAPAARSLDAAKHQQALRTWLD-------
MAV_1059|M.avium_104 VAQRTAMINRLRWRVHELDPERAPGPASLDRGKHRHLLGAWLA-------
Mflv_0450|M.gilvum_PYR-GCK VAQRTSTINRLRQRVHELDPAAEPNPGSLHRPTPCATLAAWLN-------
TH_3017|M.thermoresistible__bu VADRTRTINRLRAQLLEYFPALERAFDYSTSKAALLLLTGYQTPDGLRRA
Mvan_0416|M.vanbaalenii_PYR-1 VADRTRTINRLRQQLVAVCPALERAAQPSQDRGWVILLARYQRPKAIRQS
:* * : : :
MMAR_3792|M.marinum_M ----------------------------NQKPSSTTAHAVARREARRLAK
MSMEG_6023|M.smegmatis_MC2_155 ----------------------------TAHTDPTAVAVLTALKALAYRA
Mb0820|M.bovis_AF2122/97 ----------------------------TQPG-------LVAELARAELT
Rv0797|M.tuberculosis_H37Rv ----------------------------TQPG-------LVAELARAELT
MAV_1059|M.avium_104 ----------------------------TVPG-------LVAELARDELA
Mflv_0450|M.gilvum_PYR-GCK ----------------------------TQTG-------ILSELARDELA
TH_3017|M.thermoresistible__bu GAARLAVWLGKRKARNADAVAAKALQAAHAQHTVIPGQQLAAAMVARLAK
Mvan_0416|M.vanbaalenii_PYR-1 GVSRLTKVLTDAGVRNAASIAAAAVAAVKTQTMRLPGEEVAAGLIADLAR
:
MMAR_3792|M.marinum_M AVLEQTVQLDDNHRQLRGLAG--QAAPGLQELPGIGP-VTAAIIICAYSH
MSMEG_6023|M.smegmatis_MC2_155 QFLHH--QEHELTEQIHTLTQ--QMNPGLRAAHGVGPDTAAALLITAGMN
Mb0820|M.bovis_AF2122/97 DIIRLTGEINTLAQRISARVH--QVAPALLEIPGCAE-LTAAKIVGEAAG
Rv0797|M.tuberculosis_H37Rv DIIRLTGEINTLAQRISARVH--QVAPALLEIPGCAE-LTAAKIVGEAAG
MAV_1059|M.avium_104 DITRLTEAINALAKRIGSRVR--TVAPALLAMPGCAE-LTAAKLVGETAG
Mflv_0450|M.gilvum_PYR-GCK DIVRLSEAINAVAARIGERVR--EVAPTLLALPGCGE-LTAAKILGETAT
TH_3017|M.thermoresistible__bu EVMALDTEIGDTDAMIEERFHRHRHAEIIVSMSGFGVTLGAEFLAATGGD
Mvan_0416|M.vanbaalenii_PYR-1 EVIALDDRIKTTDADIEGRFRRHPLAEVITSMPGIGFRLGAEFLAAVG-D
. : : * . * :
MMAR_3792|M.marinum_M RGRIRSEAAFAALGGVAPLPASSGNTS-RHRLSRAGDRQLNRAFEVIVRT
MSMEG_6023|M.smegmatis_MC2_155 PHRLRSEAAFAALCGAAPVPASSGKTT-RHRLSRGGDRTANNALYRIALV
Mb0820|M.bovis_AF2122/97 VTRFKSEAAFACHAAVAPIPVWSGNTAGQMRLSRSGNRQLNAALHRIALT
Rv0797|M.tuberculosis_H37Rv VTRFKSEAAFACHAAVAPIPVWSGNTAGQMRLSRSGNRQLNAALHRIALT
MAV_1059|M.avium_104 VTRFKSEAAFARHAGVAPVPVWSGNTAGRVRMTRSGNRQLNAALHRIAIT
Mflv_0450|M.gilvum_PYR-GCK VSRFRSEAAFARHTGTAPIPVWSGNTAGRVRLSRSGNRQLNAALHRIAIT
TH_3017|M.thermoresistible__bu MSAFDSVDRLAGVAGLAPVPRDSGRISGNLQRPRRYNRRLLRVCYLSALS
Mvan_0416|M.vanbaalenii_PYR-1 PALIGSADQLAAWAGLAPVSRDSGKRTGRLHTPKRYSRRLRRVMYMSALT
: * :* . **:. **. : . : .: .* . .
MMAR_3792|M.marinum_M RMTCDP-ATRHYVARRTAEGMSKRETRRCLKRYVCRSVFRHLQAATT---
MSMEG_6023|M.smegmatis_MC2_155 RMSNDP-RTPDYVARQTANGRSKIEIIRLLKRAVAREVFRLLSQPCAIDD
Mb0820|M.bovis_AF2122/97 QIRMTDSRGQAYYQRLQDAGKTKRAALRCLKRRLARTVFQALRTVHQPSS
Rv0797|M.tuberculosis_H37Rv QIRMTDSRGQAYYQRLQDAGKTKRAALRCLKRRLARTVFQALRTVHQPSS
MAV_1059|M.avium_104 QIRLEG-LGQTYYRHRLAAGDSNPEALRCLKRRLARIIFGHLHTDHQHRT
Mflv_0450|M.gilvum_PYR-GCK QIRLTDSAGQRYYRKRLAEGKATKEALRCLKRQLARIVYRHLHTDENTRK
TH_3017|M.thermoresistible__bu SIRSDP-ASRTYYNRKRAEGKRHSQAVLALARRRLNVLWAMLRDHTTYQP
Mvan_0416|M.vanbaalenii_PYR-1 AIRCDP-DSKAYYQRKRDEGKRPIPATICLARRRTNVLYALIRDNRTWQP
: * : * * * . :: :
MMAR_3792|M.marinum_M --------------------------------------------------
MSMEG_6023|M.smegmatis_MC2_155 YSDLRPARQAKNITLTTVANHLDVWPNDISRLERGLKRDDTLAANYREYL
Mb0820|M.bovis_AF2122/97 EHTQPAAACHRSYCSRSCLSG-----------------------------
Rv0797|M.tuberculosis_H37Rv EHTQPAAACHRSYCSRSCLSG-----------------------------
MAV_1059|M.avium_104 QPCQTAAA------------------------------------------
Mflv_0450|M.gilvum_PYR-GCK TKPHNNAQTAA---------------------------------------
TH_3017|M.thermoresistible__bu TTPTAAAAA-----------------------------------------
Mvan_0416|M.vanbaalenii_PYR-1 DSPPVTAAA-----------------------------------------
MMAR_3792|M.marinum_M ---
MSMEG_6023|M.smegmatis_MC2_155 SAA
Mb0820|M.bovis_AF2122/97 ---
Rv0797|M.tuberculosis_H37Rv ---
MAV_1059|M.avium_104 ---
Mflv_0450|M.gilvum_PYR-GCK ---
TH_3017|M.thermoresistible__bu ---
Mvan_0416|M.vanbaalenii_PYR-1 ---