For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MTHAIRPVDFDNLKTMTYEVTGRIARITFNRPEKGNAIIADTPLELSALVERADLDPGVHVILVSGRGEG FCAGFDLSAYAEGSSSTGGGGAYQGTVLDGKTQAVNHLPNQPWDPMIDYQMMSRFVRGFASLMHADKPTV VKIHGYCVAGGTDIALHADQVIAAADAKIGYPPTRVWGVPAAGLWAHRLGDQRAKRLLFTGDCITGAQAA EWGLAVEAPEPADLDERTERLVARIAALPVNQLIMVKLALNSALLQQGVATSRMVSTVFDGAARHTPEGH AFVADAVEHGFRDAVRRRDEPFGDYGRQASRV
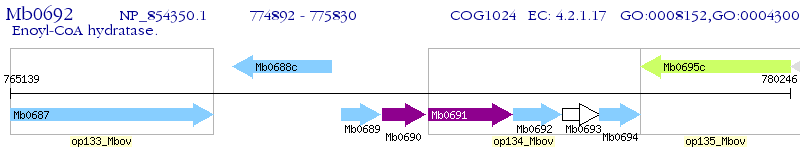
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0692 | echA4 | - | 100% (312) | enoyl-CoA hydratase |
| M. bovis AF2122 / 97 | Mb0464c | echA2 | 3e-48 | 35.76% (302) | enoyl-CoA hydratase |
| M. bovis AF2122 / 97 | Mb0694 | echA5 | 7e-20 | 30.00% (280) | enoyl-CoA hydratase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. gilvum PYR-GCK | Mflv_2433 | - | 5e-22 | 30.45% (266) | enoyl-CoA hydratase |
| M. tuberculosis H37Rv | Rv0673 | echA4 | 0.0 | 100.00% (312) | enoyl-CoA hydratase |
| M. leprae Br4923 | MLBr_02402 | - | 3e-14 | 23.94% (284) | enoyl-CoA hydratase |
| M. abscessus ATCC 19977 | MAB_3859c | - | 1e-138 | 76.16% (302) | enoyl-CoA hydratase |
| M. marinum M | MMAR_1002 | echA4 | 1e-170 | 92.63% (312) | enoyl-CoA hydratase, EchA4 |
| M. avium 104 | MAV_4499 | - | 1e-170 | 92.63% (312) | enoyl-CoA hydratase |
| M. smegmatis MC2 155 | MSMEG_1388 | - | 1e-165 | 89.74% (312) | enoyl-CoA hydratase |
| M. thermoresistible (build 8) | TH_1117 | - | 2e-22 | 30.07% (286) | enoyl-CoA hydratase/isomerase family protein |
| M. ulcerans Agy99 | MUL_0754 | echA4 | 1e-168 | 91.67% (312) | enoyl-CoA hydratase |
| M. vanbaalenii PYR-1 | Mvan_1270 | - | 1e-151 | 86.42% (302) | enoyl-CoA hydratase |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0692|M.bovis_AF2122/97 MTHAIRPVDFDNLKTMTYEVTGRIARITFNRPEKGNAIIADTPLELSALV
Rv0673|M.tuberculosis_H37Rv MTHAIRPVDFDNLKTMTYEVTGRIARITFNRPEKGNAIIADTPLELSALV
MMAR_1002|M.marinum_M MTHAIRPVDFDNLKTMTYEVTDRVARITFNRPEKGNAIVADTPLELSALV
MUL_0754|M.ulcerans_Agy99 MTHAIRPVDFDNLKTMTYEVTDRVARITFNRPEKGNAIVADTPLELSALV
MAV_4499|M.avium_104 MTHAIRPVDFDNLKTMTYEVTDRVARITFNRPEKGNAIVADTPLELSALV
MSMEG_1388|M.smegmatis_MC2_155 MTHAIRPVDFDNLKTMTYEVTDRVARITFNRPEKGNAIVADTPLELSALV
Mvan_1270|M.vanbaalenii_PYR-1 MT--------ETLKTMTYEVTGRVARITFNRPETGNAIIADTPLELAALV
MAB_3859c|M.abscessus_ATCC_199 --------MVDTLKTMTYEVTDRVARITFNRPEQGNAIIADTPLELAALV
Mflv_2433|M.gilvum_PYR-GCK -------------MYIDYEVSDRIATITLNRPEAANAQNPELLDELDAAW
TH_1117|M.thermoresistible__bu ------MPTTDDRVLFDVDRDKRIATITLNNPAQRNSYDAAMRDALERCL
MLBr_02402|M.leprae_Br4923 ----------MKYDTILVDGYQRVGIITLNRPQALNALNSQMMNEITNAA
: : *:. **:*.* *: . :
Mb0692|M.bovis_AF2122/97 ERADLDPGVHVILVSGRGEGFCAGFDLSAYAEGSSSTGGGGAYQGTVLDG
Rv0673|M.tuberculosis_H37Rv ERADLDPGVHVILVSGRGEGFCAGFDLSAYAEGSSSTGGGGAYQGTVLDG
MMAR_1002|M.marinum_M ERADLDPAVHVILVSGRGEGFCAGFDLSAYAEGSSSSGAEGGYRGTVLDG
MUL_0754|M.ulcerans_Agy99 ERADLDPAVQVILVSGRGEGFCAGFDLSAYAEGSSSSGAEGGYRGTVLDG
MAV_4499|M.avium_104 ERADLDPNVHVILVSGRGAGFCAGFDLSAYADRTGSAGGTGAYAGTVLDG
MSMEG_1388|M.smegmatis_MC2_155 ERADLDPDVHVILVSGRGEGFCAGFDLSAYAEGSSSAGGGSPYEGTVLSG
Mvan_1270|M.vanbaalenii_PYR-1 ERADLDPSVHVILVSGRGEGFCAGFDLGAYAEGSSTAGTDR--RGTVLDG
MAB_3859c|M.abscessus_ATCC_199 ERADLDPQVHVILVSGRGDGFCGGFDLSAYAEANDE--GSVRYSGTALDG
Mflv_2433|M.gilvum_PYR-GCK TRAAEDNDVSVIVLRANGKHFSAGHDLRGGGP------------------
TH_1117|M.thermoresistible__bu DQVAEDDDLTVVLLRGADGVFSTGADMNNAYAWYGE--------------
MLBr_02402|M.leprae_Br4923 KELDIDPDVGAILITGSPKVFAAGADIK----------------------
. * : .::: . *. * *:
Mb0692|M.bovis_AF2122/97 KTQAVNHLPNQPWDPMIDYQMMSRFVRGFASLMHADKPTVVKIHGYCVAG
Rv0673|M.tuberculosis_H37Rv KTQAVNHLPNQPWDPMIDYQMMSRFVRGFASLMHADKPTVVKIHGYCVAG
MMAR_1002|M.marinum_M KAQAVNHLPNQPWDPMIDYQMMSRFVRGFSSLMHADKPTVVKIHGYCVAG
MUL_0754|M.ulcerans_Agy99 KAQAVNHLPNQPWDPMIDYQMMSRFVRGFSSLTHADKPTVVKIHGYCVAG
MAV_4499|M.avium_104 KTQAVNHLPDQPWDPMIDYQMMSRFVRGFSSLMHADKPTVVKIHGYCVAG
MSMEG_1388|M.smegmatis_MC2_155 KTQALNHLPDEPWDPMVDYQMMSRFVRGFASLMHCDKPTVVKIHGYCVAG
Mvan_1270|M.vanbaalenii_PYR-1 KTQAINHRADQPWDPMIDYQMMSRFVRGFSSLMRCDKPTVVKIHGYCVAG
MAB_3859c|M.abscessus_ATCC_199 QVQLRNHLPNINWDPMLDYQMMSRFTRGFSSLLHANKPTVVKVHGYCVAG
Mflv_2433|M.gilvum_PYR-GCK -------VPDKITLEFIIQHEAKRYLEYTLRWRNVPKPSIAAVQGRCISG
TH_1117|M.thermoresistible__bu ---ENSPQARRRPSQRRRLTVDRRTFGFYHNFMGFPKVTVGEISGYALGG
MLBr_02402|M.leprae_Br4923 ----------EMASLTFTDAFDADFFSAWGKLAAVRTPMIAAVAGYALGG
. : : * .:.*
Mb0692|M.bovis_AF2122/97 GTDIALHADQVIAAADAKIGYPPTRVWGVPAAGLWAH---RLGDQRAKRL
Rv0673|M.tuberculosis_H37Rv GTDIALHADQVIAAADAKIGYPPTRVWGVPAAGLWAH---RLGDQRAKRL
MMAR_1002|M.marinum_M GTDIALHADQVIAAADAKIGYPPTRVWGVPAAGLWAH---RLGDQRAKRL
MUL_0754|M.ulcerans_Agy99 GTDIALHADQVIAAADAKIGYPPTRVWGVPAAGLWAH---RLGDQRAKRL
MAV_4499|M.avium_104 GTDIALHADQVIAAADAKIGYPPTRVWGVPAAGLWAH---RLGDQRAKRL
MSMEG_1388|M.smegmatis_MC2_155 GTDIALHADQVIAAADAKIGYPPMRVWGVPAAGLWAH---RLGDQRAKRL
Mvan_1270|M.vanbaalenii_PYR-1 GTDIALHADQVIAAADAKIGYPPTRVWGVPAAGMWAH---RLGDQRAKRL
MAB_3859c|M.abscessus_ATCC_199 GTDIALHADQLIVAADAKIGYPPTRVWGVPATGLWAH---KLGDQRAKRL
Mflv_2433|M.gilvum_PYR-GCK GLLLCWPCDLIVAADDAQFSDP-VVLMGIGGVEYHGHTW-ELGPRKAKEI
TH_1117|M.thermoresistible__bu GFELALMCDIAVIGRDAKIGMPATRFLGPALGSLHMFFH-RLGPVLARRL
MLBr_02402|M.leprae_Br4923 GCELAMMCDLLIAADTAKFGQPEIKLGVLPGMGGSQRLTRAIGKAKAMDL
* :. .* : . *::. * . :* * :
Mb0692|M.bovis_AF2122/97 LFTGDCITGAQAAEWGLAVEAPEPADLDERTERLVARIAALPVNQLIMVK
Rv0673|M.tuberculosis_H37Rv LFTGDCITGAQAAEWGLAVEAPEPADLDERTERLVARIAALPVNQLIMVK
MMAR_1002|M.marinum_M LFTGDCITGAQAAEWGLAVEAPDPKDLDERTERLVERIAALPVNQLIMIK
MUL_0754|M.ulcerans_Agy99 LFTGDCITGAQAAEWGLAVEAPDPKDLDERTERLVERIAALPVNQLIMIK
MAV_4499|M.avium_104 LFTGDCITGAQAAEWGLAVEAPDPADLDERTERLVQRIAAVPVNQLIMVK
MSMEG_1388|M.smegmatis_MC2_155 LFTGDCITGAQAAEWGLAVEAPDPADLDARTERLVERIAAMPVNQLIMAK
Mvan_1270|M.vanbaalenii_PYR-1 LLTGDCLTGAQAAEWGLAVEAPDPADLDERTERLVERIAAVPVNQLIMVK
MAB_3859c|M.abscessus_ATCC_199 LLTGDCLSGRQAYDWGLAVEAPEPDELDERTERLVERIAAMPLNQLMMIK
Mflv_2433|M.gilvum_PYR-GCK LFTGRAMTADEVAATGMVNKVVPRAELDAETRALAEQIAKMPPFALRQAK
TH_1117|M.thermoresistible__bu LLTGDIIEAGSVEHLGVFTETCDPAMVGPRARYWAEKAAKMPADGVVIAK
MLBr_02402|M.leprae_Br4923 ILTGRTIDAAEAERSGLVSRVVLADDLLPEAKAVATTISQMSRSATRMAK
::** : . .. *: .. : .:. . : :. *
Mb0692|M.bovis_AF2122/97 LALN---SALLQQGVATSRMVSTVFDGAARHTPEGHAFVADAVEHGFRDA
Rv0673|M.tuberculosis_H37Rv LALN---SALLQQGVATSRMVSTVFDGAARHTPEGHAFVADAVEHGFRDA
MMAR_1002|M.marinum_M LALN---SALLQQGVVNSRMLSTVFDGVARHTPEGHAFVADAVEHGFRDA
MUL_0754|M.ulcerans_Agy99 LALN---SALLQQGGVNSRMLSTVFDGVARHTPEGHAFVADAVEHGFRDA
MAV_4499|M.avium_104 LALN---SALLQQGVATSRMVSTVFDGVARHTPEGHAFVADAVKHGFREA
MSMEG_1388|M.smegmatis_MC2_155 LACN---TALLNQGVATSQMVSTVFDGIARHTPEGHAFVATAREHGFREA
Mvan_1270|M.vanbaalenii_PYR-1 LAMN---SALYNQGVATSAMISTVFDGIARHTPEGHAFVAQAREHGFREA
MAB_3859c|M.abscessus_ATCC_199 LACN---SVLINQGIANSAMIGTVFDGISRHTPEAHAFTADAIANGYRQA
Mflv_2433|M.gilvum_PYR-GCK RAVN---QTLDVQG--FYAAIQSVFD--VHQTGHGNALSVGGWPVLVNLE
TH_1117|M.thermoresistible__bu EAFRLVEQSQAYQGEEVASYLFHAFGTNLQFAPGEFNFVKTRAEHGTKEA
MLBr_02402|M.leprae_Br4923 EAVNRSFESTLAEGLLHERRLFHSTFVTDDQSEGMAAFIEKRAPQFTHR-
* . :* : : : .
Mb0692|M.bovis_AF2122/97 VRRRDEPFGDYGRQASRV
Rv0673|M.tuberculosis_H37Rv VRRRDEPFGDYGRQASRV
MMAR_1002|M.marinum_M VKHRDEPFGDYGRRASSV
MUL_0754|M.ulcerans_Agy99 VNHRDEPFGDYGRRASSV
MAV_4499|M.avium_104 VRHRDEPFGDHGRQASQV
MSMEG_1388|M.smegmatis_MC2_155 VRRRDEPMGDHGRRASDV
Mvan_1270|M.vanbaalenii_PYR-1 VRGRDEPFGDYGRTTSGV
MAB_3859c|M.abscessus_ATCC_199 VRHRDEPFGDYGRKPTEL
Mflv_2433|M.gilvum_PYR-GCK EMKANIK-----------
TH_1117|M.thermoresistible__bu FRLRDEHFHVPEPEPR--
MLBr_02402|M.leprae_Br4923 ------------------