For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
TELRFDDRVAVVTGAGRGLGRAYALLLAARGAKVAVNDTGGSLAGAGGDPAPAHQVVAEITALGGQAAAS LDSVTTPAGGQAIIETALEHYGRIDILIHNAGNVRSAPLKEMSYEDFDAVIDVHLRGAFHVVRPAFGLMC DQLYGRIVLASSIGGLYGNQGVANYAAAKAGLIGLSNVAAVEGAASGVLCNVIVPSAVTRMADGLDISSY PPMGTELVAPVVGLLAHESCPVNGEMLIAIAGRVARAVVAESPGVQRPSWSMEDVAEGLYAIRDLAAPLV FPVVPSGHSDHIRYSFESAAWANRGSAHHV*
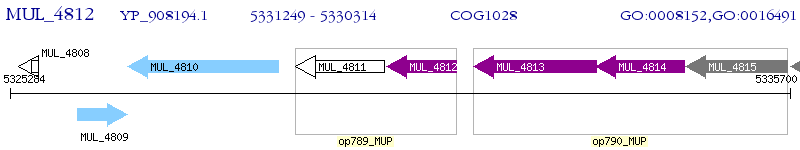
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_4812 | - | - | 100% (311) | short-chain type dehydrogenase/reductase |
| M. ulcerans Agy99 | MUL_4759 | - | 3e-56 | 45.79% (273) | short-chain type dehydrogenase/reductase |
| M. ulcerans Agy99 | MUL_4110 | - | 2e-41 | 39.93% (288) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0153 | - | 4e-56 | 45.42% (273) | short-chain type dehydrogenase/reductase |
| M. gilvum PYR-GCK | Mflv_1790 | - | 1e-122 | 71.33% (300) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv0148 | - | 4e-56 | 45.42% (273) | short-chain type dehydrogenase/reductase |
| M. leprae Br4923 | MLBr_01807 | fabG1 | 8e-22 | 32.00% (250) | 3-oxoacyl- |
| M. abscessus ATCC 19977 | MAB_3440c | - | 3e-56 | 45.07% (284) | putative short-chain dehydrogenase/reductase |
| M. marinum M | MMAR_0310 | - | 1e-173 | 99.04% (311) | short-chain type dehydrogenase/reductase |
| M. avium 104 | MAV_5205 | - | 1e-128 | 76.08% (301) | oxidoreductase, short chain dehydrogenase/reductase family |
| M. smegmatis MC2 155 | MSMEG_5624 | - | 1e-123 | 70.43% (301) | oxidoreductase, short chain dehydrogenase/reductase family |
| M. thermoresistible (build 8) | TH_2730 | - | 1e-79 | 70.79% (202) | PUTATIVE short-chain dehydrogenase/reductase SDR |
| M. vanbaalenii PYR-1 | Mvan_4960 | - | 1e-116 | 69.16% (308) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
Mb0153|M.bovis_AF2122/97 MPG----VQDRVIVVTGAGGGLGREYALTLAGEGASVVVNDLGGARDGTG
Rv0148|M.tuberculosis_H37Rv MPG----VQDRVIVVTGAGGGLGREYALTLAGEGASVVVNDLGGARDGTG
MAB_3440c|M.abscessus_ATCC_199 MFGTDVRFDGRVAIVTGAGGGLGREYALLLASRGAKVVVNDIGSSDELLG
MUL_4812|M.ulcerans_Agy99 --MTELRFDDRVAVVTGAGRGLGRAYALLLAARGAKVAVNDTGGS--LAG
MMAR_0310|M.marinum_M --MTELRFDDRVAVVTGAGRGLGRAYALLLAARGAKVVVNDTGGS--LAG
MAV_5205|M.avium_104 -----------MAVVTGAGRGLGRAYAHLLAARGAKVVVNDVGGA--LDG
Mflv_1790|M.gilvum_PYR-GCK --MTDLRFDGRVAAVTGGGRGLGRAYALMLASRGAKVVVNDVGGS--LTG
TH_2730|M.thermoresistible__bu --------------------------------------VRSAG-------
Mvan_4960|M.vanbaalenii_PYR-1 --MADLRFDGRVAVVTGAGRGLGRAYALMLAADGAKVVVNDSGGA--LDG
MSMEG_5624|M.smegmatis_MC2_155 --MSALRFDDRVAVVTGAGRGLGREYALLLAARGAKVVVNDAGVS--LTG
MLBr_01807|M.leprae_Br4923 ------MTDAAAKVSAAPGRGK-----PAFVSRSILVTGGNRGIGLAVAR
. *
Mb0153|M.bovis_AF2122/97 AGS--AMADEVVAEIRDKGGRAVANYDSVATEDGAANIIKTALDEFGAVH
Rv0148|M.tuberculosis_H37Rv AGS--AMADEVVAEIRDKGGRAVANYDSVATEDGAANIIKTALDEFGAVH
MAB_3440c|M.abscessus_ATCC_199 GGSSDAPANSVVAEIQALGGDAIADTNTVATGAGGRAIVGTALSAWGRVD
MUL_4812|M.ulcerans_Agy99 AGGDPAPAHQVVAEITALGGQAAASLDSVTTPAGGQAIIETALEHYGRID
MMAR_0310|M.marinum_M AGGDPAPAHQVVAEITALGGQAAASLDSVTTPAGGQAIIETALERYGRID
MAV_5205|M.avium_104 AGVDTGPAAQVVDEITAAGGDAVACTESVATPEGGRAIIETALAHYGRLD
Mflv_1790|M.gilvum_PYR-GCK DGADPGPAQQVADEIVAAGGEAVANTDSVATADGGAAIVATALDAYGRID
TH_2730|M.thermoresistible__bu ----P-PAEELGXXX-----------------------------------
Mvan_4960|M.vanbaalenii_PYR-1 EGADTGPAQAVVDEIASAGGEAVANTDSVATPEGGRAIIADAQQRFGGVD
MSMEG_5624|M.smegmatis_MC2_155 DGSDEGPAHAVVEEIVAAGGRAVASTDSVATAAGGRAIVDTALEHYGRVD
MLBr_01807|M.leprae_Br4923 RLVADGHRVAVTHRASVVPDWFFGVECDVTDNDAIGAAFKAVEEHQGPVE
:
Mb0153|M.bovis_AF2122/97 GVVSNAGILR-DGTFHKMSFENWDAVLKVHLYGGYHVLRAAWPHFREQSY
Rv0148|M.tuberculosis_H37Rv GVVSNAGILR-DGTFHKMSFENWDAVLKVHLYGGYHVLRAAWPHFREQSY
MAB_3440c|M.abscessus_ATCC_199 IVINNAGIVGPIKTFAELTDEDVDTVFGVQLLGPFNVLRPAWKAMVDQGY
MUL_4812|M.ulcerans_Agy99 ILIHNAGNVR-SAPLKEMSYEDFDAVIDVHLRGAFHVVRPAFGLMCDQLY
MMAR_0310|M.marinum_M ILIHNAGNVR-SAPLKEMSYEDFDAVIDVHLRGAFHVVRPAFGLMCDQLY
MAV_5205|M.avium_104 VLVHNAGNVR-RASLKQMSYEDFDAVLDVHLRGAFHVLRPAFPVMCRAGY
Mflv_1790|M.gilvum_PYR-GCK VLVHNAGNVR-RGSLREMSHDDFDAVLDVHLRGAFHVVRPAFPVMCDNGY
TH_2730|M.thermoresistible__bu --IHNAGTVR-RGSLRDLSQDDFDAVLDVHLRGAFHVLRPAFPVMCDAGY
Mvan_4960|M.vanbaalenii_PYR-1 ILIHNAGIVR-RAPLAEMTYDDFEAVLDVHLRGAFHVVRPAFPSMCDAGY
MSMEG_5624|M.smegmatis_MC2_155 ILIHNAGNVR-RGSLRELGDDDFDAVLDVHLRGAYHVVRPAFPVMCDAGY
MLBr_01807|M.leprae_Br4923 VLVANAGITS-DMLIMRMSEEQFQKVIDVNLTGVFRVAKLASRNMIKQRF
: *** : : :: : *: *:* * :.* : * : :
Mb0153|M.bovis_AF2122/97 GRVVVATSTSGLFGNFGQTNYGAAKLGLVGLINTLALEGAKYNIHANALA
Rv0148|M.tuberculosis_H37Rv GRVVVATSTSGLFGNFGQTNYGAAKLGLVGLINTLALEGAKYNIHANALA
MAB_3440c|M.abscessus_ATCC_199 GRILNISSGS-IFGQGEACTYPTAKAGMVGMTTNLGLAGPAYGIKVNALL
MUL_4812|M.ulcerans_Agy99 GRIVLASSIGGLYGNQGVANYAAAKAGLIGLSNVAAVEGAASGVLCNVIV
MMAR_0310|M.marinum_M GRIVLASSIGGLYGNQGVANYAAAKAGLIGLSNVAAVEGAASGVLCNVIV
MAV_5205|M.avium_104 GRIVLTSSIGGLYGNQGVANYAAAKAGVIGLSNVAALEGAAEGVRCNVIV
Mflv_1790|M.gilvum_PYR-GCK GRIVLTSSIGGLYGNHDVANYAVAKAGVIGLSNVAALEGAAHGVRSNVIV
TH_2730|M.thermoresistible__bu GRIVLTSSIGGLYGNHEVANYAVAKSGVIGLSNVAALEGAEFGVKSNVVV
Mvan_4960|M.vanbaalenii_PYR-1 GRIVLTSSIGGLYGNHEVANYAVAKAGVIGLSHVAAMEGAAHDVKSNVIV
MSMEG_5624|M.smegmatis_MC2_155 GRVVLTSSIGGLYGNHGVVNYAAAKAGILGLCNVVALEGAADGVTCNAII
MLBr_01807|M.leprae_Br4923 GRYIFMGSVVGLSGVPGQANYAASKSGLIGMARSLARELGSRSITANVVA
** : * : * .* .:* *::*: . .: *.:
Mb0153|M.bovis_AF2122/97 PIAATRMTQDILPPEV---LEKLTPEFVAPVVAYLCTEECADNASVYVVG
Rv0148|M.tuberculosis_H37Rv PIAATRMTQDILPPEV---LEKLTPEFVAPVVAYLCTEECADNASVYVVG
MAB_3440c|M.abscessus_ATCC_199 PVGFTRMVENMPEPALSWLRDTFPAALVAPVAAYLVSEDVPCSGLSFSAG
MUL_4812|M.ulcerans_Agy99 PSAVTRMADGLDISSYP----PMGTELVAPVVGLLAHESCPVNGEMLIAI
MMAR_0310|M.marinum_M PSAVTRMADGLDISSYP----PMGTELVAPVVGLLAHESCPVNGEMLIAI
MAV_5205|M.avium_104 PAALTRMADGIDTSAYP----PMGPELVAPVVGWLAHESCSVSGELFIAL
Mflv_1790|M.gilvum_PYR-GCK PAAVTRMAEGIDTSAYP----PMDPDLVAPTVGWLAHETCSVTGEIFIAL
TH_2730|M.thermoresistible__bu PAALTRMAEGIDTSAYP----PMGPELVAPLVGWLAHESCSVTGEMLIGI
Mvan_4960|M.vanbaalenii_PYR-1 PAAVTRMAAGIDTSAYP----PMGPELVAPVVGYLAHETCAVTGEMFVAL
MSMEG_5624|M.smegmatis_MC2_155 PGAVTRMAEGLDTSAYP----PMGPDLVAPAVGWLAHETCSVTGEYLVAI
MLBr_01807|M.leprae_Br4923 PGFIDTEMTRAMDSKYQ------EAALQAIPLGRIGAVEEIAGAVSFLAS
* . . : * . : .
Mb0153|M.bovis_AF2122/97 GGKVQRVALFGNDGANFDKPPSVQDVAARWAEITDLSGAKIAG-------
Rv0148|M.tuberculosis_H37Rv GGKVQRVALFGNDGANFDKPPSVQDVAARWAEITDLSGAKIAG-------
MAB_3440c|M.abscessus_ATCC_199 GGRFARVFLGEARGVTSS-QLTIEEVRDRFEAAMEIEGAATMANVGD---
MUL_4812|M.ulcerans_Agy99 AGRVARAVVAESPGVQRP-SWSMEDVAEGLYAIRDLAAPLVFPVVPSGHS
MMAR_0310|M.marinum_M AGRVARAVVAESPGVQRL-SWSMEDVAEGLYAIRDLAAPLVFPVVPSGHS
MAV_5205|M.avium_104 AGRVARAVIAESPGVCRP-GWTVEDVGEHLDAIRYVEAPLIFPVVPDGHA
Mflv_1790|M.gilvum_PYR-GCK AGRVARAVIAETPGVYQP-SWTIEDVATRIDEIRDFSAPVVFPVVPSGHN
TH_2730|M.thermoresistible__bu AGRMARAAVVESPGVYRG-SWTIEDVADQLEAIRDLGDPVVFDVVPTGHN
Mvan_4960|M.vanbaalenii_PYR-1 AGRVATAAVTETRGVYRP-SWTIADVAENLDAIRDRSESLTFPVVPNGHG
MSMEG_5624|M.smegmatis_MC2_155 AGRIGRAALVESPGVYQP-SWTVEQVGERIDEIRDISAPVTFPVVPDGHA
MLBr_01807|M.leprae_Br4923 -----------------------EDAGYISGAVIPVDGGLGMGH------
:.
Mb0153|M.bovis_AF2122/97 ----FKL------------------
Rv0148|M.tuberculosis_H37Rv ----FKL------------------
MAB_3440c|M.abscessus_ATCC_199 ---ELRLYAAALTGR----------
MUL_4812|M.ulcerans_Agy99 DHIRYSFESAAWANRGSAHHV----
MMAR_0310|M.marinum_M DHIRYSFESAAWANRGSAHHV----
MAV_5205|M.avium_104 EHIRYSFELAQRANEQGALHG----
Mflv_1790|M.gilvum_PYR-GCK DHIGYSFAMADERSREERVASHG--
TH_2730|M.thermoresistible__bu EHIGYSFAMARQEAHSG--------
Mvan_4960|M.vanbaalenii_PYR-1 DHIRYSFAMASRAAAMSACAKSRSR
MSMEG_5624|M.smegmatis_MC2_155 EHIRYSFAQAAQGVGS---------
MLBr_01807|M.leprae_Br4923 -------------------------