For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
FGTDVRFDGRVAIVTGAGGGLGREYALLLASRGAKVVVNDIGSSDELLGGGSSDAPANSVVAEIQALGGD AIADTNTVATGAGGRAIVGTALSAWGRVDIVINNAGIVGPIKTFAELTDEDVDTVFGVQLLGPFNVLRPA WKAMVDQGYGRILNISSGSIFGQGEACTYPTAKAGMVGMTTNLGLAGPAYGIKVNALLPVGFTRMVENMP EPALSWLRDTFPAALVAPVAAYLVSEDVPCSGLSFSAGGGRFARVFLGEARGVTSSQLTIEEVRDRFEAA MEIEGAATMANVGDELRLYAAALTGR*
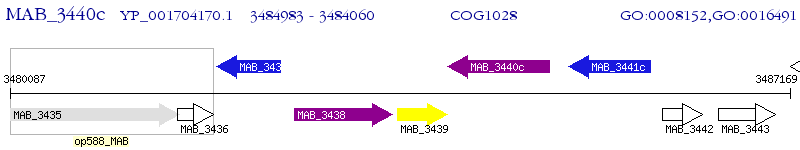
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. abscessus ATCC 19977 | MAB_3440c | - | - | 100% (307) | putative short-chain dehydrogenase/reductase |
| M. abscessus ATCC 19977 | MAB_4604c | - | 1e-50 | 43.01% (286) | putative short-chain dehydrogenase/reductase |
| M. abscessus ATCC 19977 | MAB_2948c | - | 2e-40 | 38.87% (265) | short chain dehydrogenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0153 | - | 9e-47 | 41.43% (280) | short-chain type dehydrogenase/reductase |
| M. gilvum PYR-GCK | Mflv_1790 | - | 2e-60 | 46.21% (277) | short-chain dehydrogenase/reductase SDR |
| M. tuberculosis H37Rv | Rv0148 | - | 9e-47 | 41.43% (280) | short-chain type dehydrogenase/reductase |
| M. leprae Br4923 | MLBr_00862 | ephD | 6e-19 | 34.87% (195) | short chain dehydrogenase |
| M. marinum M | MMAR_0310 | - | 1e-56 | 45.42% (284) | short-chain type dehydrogenase/reductase |
| M. avium 104 | MAV_5205 | - | 5e-54 | 43.51% (285) | oxidoreductase, short chain dehydrogenase/reductase family |
| M. smegmatis MC2 155 | MSMEG_5624 | - | 9e-60 | 46.74% (291) | oxidoreductase, short chain dehydrogenase/reductase family |
| M. thermoresistible (build 8) | TH_4146 | - | 9e-53 | 45.15% (268) | PUTATIVE putative short-chain dehydrogenase/reductase SDR |
| M. ulcerans Agy99 | MUL_4812 | - | 2e-56 | 45.07% (284) | short-chain type dehydrogenase/reductase |
| M. vanbaalenii PYR-1 | Mvan_4960 | - | 2e-57 | 43.51% (308) | short-chain dehydrogenase/reductase SDR |
CLUSTAL 2.0.9 multiple sequence alignment
MMAR_0310|M.marinum_M --------------------------------------------------
MUL_4812|M.ulcerans_Agy99 --------------------------------------------------
MAV_5205|M.avium_104 --------------------------------------------------
Mvan_4960|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_1790|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5624|M.smegmatis_MC2_155 --------------------------------------------------
Mb0153|M.bovis_AF2122/97 --------------------------------------------------
Rv0148|M.tuberculosis_H37Rv --------------------------------------------------
TH_4146|M.thermoresistible__bu --------------------------------------------------
MAB_3440c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 MPATQSIHKVPHQFVESPDGVRLAIYQSGQPEGPAIVLVHGFPDSHVLWD
MMAR_0310|M.marinum_M --------------------------------------------------
MUL_4812|M.ulcerans_Agy99 --------------------------------------------------
MAV_5205|M.avium_104 --------------------------------------------------
Mvan_4960|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_1790|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5624|M.smegmatis_MC2_155 --------------------------------------------------
Mb0153|M.bovis_AF2122/97 --------------------------------------------------
Rv0148|M.tuberculosis_H37Rv --------------------------------------------------
TH_4146|M.thermoresistible__bu --------------------------------------------------
MAB_3440c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 GVVPLLAKRFRIVRYDNRGVGQSSVPKPVSAYTMTRFADDFAAVIDELSP
MMAR_0310|M.marinum_M --------------------------------------------------
MUL_4812|M.ulcerans_Agy99 --------------------------------------------------
MAV_5205|M.avium_104 --------------------------------------------------
Mvan_4960|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_1790|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5624|M.smegmatis_MC2_155 --------------------------------------------------
Mb0153|M.bovis_AF2122/97 --------------------------------------------------
Rv0148|M.tuberculosis_H37Rv --------------------------------------------------
TH_4146|M.thermoresistible__bu --------------------------------------------------
MAB_3440c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DRPVHVLAHDWGSVGVWEYLSRSGASDRVASFTSVSGPSQDQLVDYILSG
MMAR_0310|M.marinum_M --------------------------------------------------
MUL_4812|M.ulcerans_Agy99 --------------------------------------------------
MAV_5205|M.avium_104 --------------------------------------------------
Mvan_4960|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_1790|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5624|M.smegmatis_MC2_155 --------------------------------------------------
Mb0153|M.bovis_AF2122/97 --------------------------------------------------
Rv0148|M.tuberculosis_H37Rv --------------------------------------------------
TH_4146|M.thermoresistible__bu --------------------------------------------------
MAB_3440c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LRRPWRPRTFARAVSQLFRLTYMVLFSIPVLVPMVLWIALSSATIRRTMV
MMAR_0310|M.marinum_M --------------------------------------------------
MUL_4812|M.ulcerans_Agy99 --------------------------------------------------
MAV_5205|M.avium_104 --------------------------------------------------
Mvan_4960|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_1790|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5624|M.smegmatis_MC2_155 --------------------------------------------------
Mb0153|M.bovis_AF2122/97 --------------------------------------------------
Rv0148|M.tuberculosis_H37Rv --------------------------------------------------
TH_4146|M.thermoresistible__bu --------------------------------------------------
MAB_3440c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 DNISTDQIHHSATLARDAAHSVKTYPANYFRAFSGRRRGKGVPVVNVPVQ
MMAR_0310|M.marinum_M --------------------------------------------------
MUL_4812|M.ulcerans_Agy99 --------------------------------------------------
MAV_5205|M.avium_104 --------------------------------------------------
Mvan_4960|M.vanbaalenii_PYR-1 --------------------------------------------------
Mflv_1790|M.gilvum_PYR-GCK --------------------------------------------------
MSMEG_5624|M.smegmatis_MC2_155 --------------------------------------------------
Mb0153|M.bovis_AF2122/97 --------------------------------------------------
Rv0148|M.tuberculosis_H37Rv --------------------------------------------------
TH_4146|M.thermoresistible__bu --------------------------------------------------
MAB_3440c|M.abscessus_ATCC_199 --------------------------------------------------
MLBr_00862|M.leprae_Br4923 LIVNTMDRYVRPYGYDATARWVPRLWRRDIKAGHFSPMSHPQVMAAAVHD
MMAR_0310|M.marinum_M --------------------MTELRFDDRVAVVTGAGRGLGRAYALLLAA
MUL_4812|M.ulcerans_Agy99 --------------------MTELRFDDRVAVVTGAGRGLGRAYALLLAA
MAV_5205|M.avium_104 -----------------------------MAVVTGAGRGLGRAYAHLLAA
Mvan_4960|M.vanbaalenii_PYR-1 --------------------MADLRFDGRVAVVTGAGRGLGRAYALMLAA
Mflv_1790|M.gilvum_PYR-GCK --------------------MTDLRFDGRVAAVTGGGRGLGRAYALMLAS
MSMEG_5624|M.smegmatis_MC2_155 --------------------MSALRFDDRVAVVTGAGRGLGREYALLLAA
Mb0153|M.bovis_AF2122/97 ----------------------MPGVQDRVIVVTGAGGGLGREYALTLAG
Rv0148|M.tuberculosis_H37Rv ----------------------MPGVQDRVIVVTGAGGGLGREYALTLAG
TH_4146|M.thermoresistible__bu ----------------------MT-FDNEVAIVTGAGRGLGRCHALELAR
MAB_3440c|M.abscessus_ATCC_199 ------------------MFGTDVRFDGRVAIVTGAGGGLGREYALLLAS
MLBr_00862|M.leprae_Br4923 LADLVDGKQPSRALLRAQVGRSRDTFGDTLVSVTGAGSGIGRETALALAR
: ***.* *:** * **
MMAR_0310|M.marinum_M RGAKVVVNDTGGS--LAGAGGDPAPAHQVVAEITALGGQAAASLDSVTTP
MUL_4812|M.ulcerans_Agy99 RGAKVAVNDTGGS--LAGAGGDPAPAHQVVAEITALGGQAAASLDSVTTP
MAV_5205|M.avium_104 RGAKVVVNDVGGA--LDGAGVDTGPAAQVVDEITAAGGDAVACTESVATP
Mvan_4960|M.vanbaalenii_PYR-1 DGAKVVVNDSGGA--LDGEGADTGPAQAVVDEIASAGGEAVANTDSVATP
Mflv_1790|M.gilvum_PYR-GCK RGAKVVVNDVGGS--LTGDGADPGPAQQVADEIVAAGGEAVANTDSVATA
MSMEG_5624|M.smegmatis_MC2_155 RGAKVVVNDAGVS--LTGDGSDEGPAHAVVEEIVAAGGRAVASTDSVATA
Mb0153|M.bovis_AF2122/97 EGASVVVNDLGGA--RDGTGAGSAMADEVVAEIRDKGGRAVANYDSVATE
Rv0148|M.tuberculosis_H37Rv EGASVVVNDLGGA--RDGTGAGSAMADEVVAEIRDKGGRAVANYDSVATE
TH_4146|M.thermoresistible__bu RGARVVVNDVGSD--VDGTGASASAAQAVVDEITAAGGTAVASTDSVATP
MAB_3440c|M.abscessus_ATCC_199 RGAKVVVNDIGSSDELLGGGSSDAPANSVVAEIQALGGDAIADTNTVATG
MLBr_00862|M.leprae_Br4923 RGAEVVVSDIDEA-----------AAKNTAAQIVASGGVAYAYALDVSDA
** *.*.* . * .. :* ** * * *:
MMAR_0310|M.marinum_M AGGQAIIETALERYGRIDILIHNAGNVR-SAPLKEMSYEDFDAVIDVHLR
MUL_4812|M.ulcerans_Agy99 AGGQAIIETALEHYGRIDILIHNAGNVR-SAPLKEMSYEDFDAVIDVHLR
MAV_5205|M.avium_104 EGGRAIIETALAHYGRLDVLVHNAGNVR-RASLKQMSYEDFDAVLDVHLR
Mvan_4960|M.vanbaalenii_PYR-1 EGGRAIIADAQQRFGGVDILIHNAGIVR-RAPLAEMTYDDFEAVLDVHLR
Mflv_1790|M.gilvum_PYR-GCK DGGAAIVATALDAYGRIDVLVHNAGNVR-RGSLREMSHDDFDAVLDVHLR
MSMEG_5624|M.smegmatis_MC2_155 AGGRAIVDTALEHYGRVDILIHNAGNVR-RGSLRELGDDDFDAVLDVHLR
Mb0153|M.bovis_AF2122/97 DGAANIIKTALDEFGAVHGVVSNAGILR-DGTFHKMSFENWDAVLKVHLY
Rv0148|M.tuberculosis_H37Rv DGAANIIKTALDEFGAVHGVVSNAGILR-DGTFHKMSFENWDAVLKVHLY
TH_4146|M.thermoresistible__bu EGGAAIVKTAMEAFGRVDILVNNAGILR-DAAFKNMTPQQVAAVLDVHLA
MAB_3440c|M.abscessus_ATCC_199 AGGRAIVGTALSAWGRVDIVINNAGIVGPIKTFAELTDEDVDTVFGVQLL
MLBr_00862|M.leprae_Br4923 AAVEAFAEQVSAKHGVPDIVVNNAGIGQ-AGRFLDTPPEEFDRVLAVNLG
. : . * . :: *** : . :: *: *:*
MMAR_0310|M.marinum_M GAFHVVRPAFGLMCDQLYG-RIVLASSIGGLYGNQGVANYAAAKAGLIGL
MUL_4812|M.ulcerans_Agy99 GAFHVVRPAFGLMCDQLYG-RIVLASSIGGLYGNQGVANYAAAKAGLIGL
MAV_5205|M.avium_104 GAFHVLRPAFPVMCRAGYG-RIVLTSSIGGLYGNQGVANYAAAKAGVIGL
Mvan_4960|M.vanbaalenii_PYR-1 GAFHVVRPAFPSMCDAGYG-RIVLTSSIGGLYGNHEVANYAVAKAGVIGL
Mflv_1790|M.gilvum_PYR-GCK GAFHVVRPAFPVMCDNGYG-RIVLTSSIGGLYGNHDVANYAVAKAGVIGL
MSMEG_5624|M.smegmatis_MC2_155 GAYHVVRPAFPVMCDAGYG-RVVLTSSIGGLYGNHGVVNYAAAKAGILGL
Mb0153|M.bovis_AF2122/97 GGYHVLRAAWPHFREQSYG-RVVVATSTSGLFGNFGQTNYGAAKLGLVGL
Rv0148|M.tuberculosis_H37Rv GGYHVLRAAWPHFREQSYG-RVVVATSTSGLFGNFGQTNYGAAKLGLVGL
TH_4146|M.thermoresistible__bu GAFHVTRAVWPIMREANHG-RIIQTSSGTGLFGNFGQANYGAAKMGLIGM
MAB_3440c|M.abscessus_ATCC_199 GPFNVLRPAWKAMVDQGYG-RILNISSGS-IFGQGEACTYPTAKAGMVGM
MLBr_00862|M.leprae_Br4923 GVVNCCRAFGQRLVERGTGGHIVNVSSMAAYAPLQSLSAYCTSKAATYMF
* : *. : * ::: :* * .:* . :
MMAR_0310|M.marinum_M SNVAAVEGAASGVLCNVIVPSAVTRMADGLDI----SSYPPMGTELVAPV
MUL_4812|M.ulcerans_Agy99 SNVAAVEGAASGVLCNVIVPSAVTRMADGLDI----SSYPPMGTELVAPV
MAV_5205|M.avium_104 SNVAALEGAAEGVRCNVIVPAALTRMADGIDT----SAYPPMGPELVAPV
Mvan_4960|M.vanbaalenii_PYR-1 SHVAAMEGAAHDVKSNVIVPAAVTRMAAGIDT----SAYPPMGPELVAPV
Mflv_1790|M.gilvum_PYR-GCK SNVAALEGAAHGVRSNVIVPAAVTRMAEGIDT----SAYPPMDPDLVAPT
MSMEG_5624|M.smegmatis_MC2_155 CNVVALEGAADGVTCNAIIPGAVTRMAEGLDT----SAYPPMGPDLVAPA
Mb0153|M.bovis_AF2122/97 INTLALEGAKYNIHANALAPIAATRMTQDILPP---EVLEKLTPEFVAPV
Rv0148|M.tuberculosis_H37Rv INTLALEGAKYNIHANALAPIAATRMTQDILPP---EVLEKLTPEFVAPV
TH_4146|M.thermoresistible__bu MHVLAIEGARDNIAVNAIAPIARTRMTEDIMG----EAGKAMDPELVTPV
MAB_3440c|M.abscessus_ATCC_199 TTNLGLAGPAYGIKVNALLPVGFTRMVENMPEPALSWLRDTFPAALVAPV
MLBr_00862|M.leprae_Br4923 SDCLRAELDAVGVGLTTICPGVIDTNIIQSTR---------IDTPTAKQE
.: ..: * : . .
MMAR_0310|M.marinum_M VGLLAHESCPVNGEMLIAIAGRVARAVVAESPGVQRL-SWSMEDVAEGLY
MUL_4812|M.ulcerans_Agy99 VGLLAHESCPVNGEMLIAIAGRVARAVVAESPGVQRP-SWSMEDVAEGLY
MAV_5205|M.avium_104 VGWLAHESCSVSGELFIALAGRVARAVIAESPGVCRP-GWTVEDVGEHLD
Mvan_4960|M.vanbaalenii_PYR-1 VGYLAHETCAVTGEMFVALAGRVATAAVTETRGVYRP-SWTIADVAENLD
Mflv_1790|M.gilvum_PYR-GCK VGWLAHETCSVTGEIFIALAGRVARAVIAETPGVYQP-SWTIEDVATRID
MSMEG_5624|M.smegmatis_MC2_155 VGWLAHETCSVTGEYLVAIAGRIGRAALVESPGVYQP-SWTVEQVGERID
Mb0153|M.bovis_AF2122/97 VAYLCTEECADNASVYVVGGGKVQRVALFGNDGANFDKPPSVQDVAARWA
Rv0148|M.tuberculosis_H37Rv VAYLCTEECADNASVYVVGGGKVQRVALFGNDGANFDKPPSVQDVAARWA
TH_4146|M.thermoresistible__bu VIYLAHRDCDRTAHIYSVGGGKVSRVFIGVSEGIAHP-ALTPESVAAAID
MAB_3440c|M.abscessus_ATCC_199 AAYLVSEDVPCSGLSFSAGGGRFARVFLGEARGVTSS-QLTIEEVRDRFE
MLBr_00862|M.leprae_Br4923 RVRDRRGQLDMMFRLRRYGPDKVADTILSAIK-KNKP----IRPVAPEAY
.:. . : *
MMAR_0310|M.marinum_M AIRDLAAPLVFPVVPSGHSDHIRYSFESAAWANRGSAHHV----
MUL_4812|M.ulcerans_Agy99 AIRDLAAPLVFPVVPSGHSDHIRYSFESAAWANRGSAHHV----
MAV_5205|M.avium_104 AIRYVEAPLIFPVVPDGHAEHIRYSFELAQRANEQGALHG----
Mvan_4960|M.vanbaalenii_PYR-1 AIRDRSESLTFPVVPNGHGDHIRYSFAMASRAAAMSACAKSRSR
Mflv_1790|M.gilvum_PYR-GCK EIRDFSAPVVFPVVPSGHNDHIGYSFAMADERSREERVASHG--
MSMEG_5624|M.smegmatis_MC2_155 EIRDISAPVTFPVVPDGHAEHIRYSFAQAAQG-----VGS----
Mb0153|M.bovis_AF2122/97 EITDLSG-----AKIAG--FKL----------------------
Rv0148|M.tuberculosis_H37Rv EITDLSG-----AKIAG--FKL----------------------
TH_4146|M.thermoresistible__bu EIDDPSS-----FTIRGGPVRVDVSTR-----------------
MAB_3440c|M.abscessus_ATCC_199 AAMEIEG----AATMANVGDELRLYAAALTGR------------
MLBr_00862|M.leprae_Br4923 ALYGISR-----VLPQVLRSTARIRVI-----------------