For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
VVECVTVIEIQAVDAVQVLTLSSGRVNALDVELLTELTDALRELGSSHGGALVLTGAGRVFSAGVDLNRV VQGGTSYTDRLIPALSDAFDAMFGYPGPTVAAINGAAIAGGCVLACACDRRLIGAEAAIGASEVLVGVAF PVVALEVMRYACGDRAEEVLLGGRSYRGIEAVSRGLAHEVVAEDLLGVATAQASELGDIPADAYRHTKAQ LRAPARARMRDGGDIDREVRQIWGSAQTQQRIAGYLERLRRR*
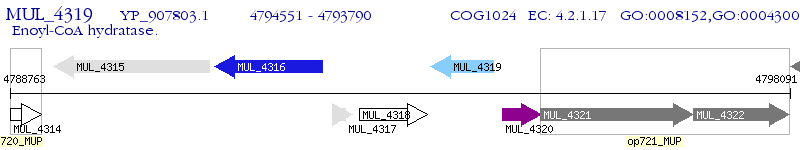
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_4319 | echA3_1 | - | 100% (253) | enoyl-CoA hydratase, EchA3_1 |
| M. ulcerans Agy99 | MUL_0713 | echA3 | 2e-20 | 30.52% (213) | enoyl-CoA hydratase |
| M. ulcerans Agy99 | MUL_4429 | echA12_2 | 3e-20 | 32.29% (223) | enoyl-CoA hydratase, EchA12_2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb0649c | echA3 | 3e-20 | 30.99% (213) | enoyl-CoA hydratase |
| M. gilvum PYR-GCK | Mflv_2036 | - | 2e-18 | 30.52% (213) | enoyl-CoA hydratase |
| M. tuberculosis H37Rv | Rv0632c | echA3 | 3e-20 | 30.99% (213) | enoyl-CoA hydratase |
| M. leprae Br4923 | MLBr_02402 | - | 1e-19 | 29.22% (219) | enoyl-CoA hydratase |
| M. abscessus ATCC 19977 | MAB_1185c | - | 5e-23 | 28.81% (243) | enoyl-CoA hydratase |
| M. marinum M | MMAR_5243 | echA3_1 | 1e-136 | 99.60% (248) | enoyl-CoA hydratase, EchA3_1 |
| M. avium 104 | MAV_4534 | - | 8e-21 | 31.28% (211) | enoyl-CoA hydratase |
| M. smegmatis MC2 155 | MSMEG_2328 | - | 2e-17 | 32.52% (206) | enoyl-CoA hydratase |
| M. thermoresistible (build 8) | TH_4102 | - | 6e-19 | 28.33% (240) | PUTATIVE short chain enoyl-CoA hydratase |
| M. vanbaalenii PYR-1 | Mvan_3990 | - | 5e-18 | 32.91% (237) | enoyl-CoA hydratase/isomerase |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_4319|M.ulcerans_Agy99 MVVECVTVIEIQAVDAVQVLTLSSGR--VNALDVELLTELTDALRELGSS
MMAR_5243|M.marinum_M -----MTVIEIQAVDAVQVLTLSSGR--VNALDVELLTELTDALRELGSS
Mflv_2036|M.gilvum_PYR-GCK ---MTFETILVDRDDRVATITLNRPK-ALNALNTQVMNEVTTAAAELDAE
TH_4102|M.thermoresistible__bu MSAKNYETILVTREDRVGIITLNRPK-ALNALNSQVMTEVTDAAAEFDSD
MAB_1185c|M.abscessus_ATCC_199 MTSHNFETILTERIDRVAVITLNRPK-ALNALNSQVMNEVTTAAAEFDAD
MLBr_02402|M.leprae_Br4923 ---MKYDTILVDGYQRVGIITLNRPQ-ALNALNSQMMNEITNAAKELDID
MSMEG_2328|M.smegmatis_MC2_155 -MNEFIKIVLGSAPEQTGVATLLLSRPPTNALTRQMYREISIAANELAQR
Mb0649c|M.bovis_AF2122/97 ----MSDPVSYTRKDSIAVISMDDGK--VNALGPAMQQALNAAIDNAD-R
Rv0632c|M.tuberculosis_H37Rv ----MSDPVSYTRKDSIAVISMDDGK--VNALGPAMQQALNAAIDNAD-R
MAV_4534|M.avium_104 ----MSGPVHYSTKGPVAVIRMDDGK--VNALGPTMQQALGEAIDRAE-A
Mvan_3990|M.vanbaalenii_PYR-1 --MSDALVVSTRPVDHVAVVMLNRPE-TLNALDRGLMDELQVSLQALAGQ
: : . *** : : :
MUL_4319|M.ulcerans_Agy99 HGG-ALVLTGAGRVFSAGVDLNRVVQGG-TSYTDRLIPALSDAFDAMFGY
MMAR_5243|M.marinum_M HGG-ALVLTGAGRVFSAGVDLNRVVQGG-TSYTDRLIPALSDAFDAMFGY
Mflv_2036|M.gilvum_PYR-GCK PGVGAIIVTGSERAFAAGADIKEMASLS-FADVFSSDFFAA--WGKFAAT
TH_4102|M.thermoresistible__bu PGIGAIIVTGNEKAFAAGADIKEMADLE-FADVFAADFFAP--WSKFAAT
MAB_1185c|M.abscessus_ATCC_199 HGIGAIIITGSEKAFAAGADIKEMSEQS-FSDMFGSDFFSA--WGKLGAV
MLBr_02402|M.leprae_Br4923 PDVGAILITGSPKVFAAGADIKEMASLT-FTDAFDADFFSA--WGKLAAV
MSMEG_2328|M.smegmatis_MC2_155 ADVSSVIVYGGHEIFCAGDDIPELRTLD-AEETAAADHALQRCIEAVAAI
Mb0649c|M.bovis_AF2122/97 DDVGALVITGNGRVFSGGFDLKILTSGE-VQPAIDMLRGGFELAYRLLSY
Rv0632c|M.tuberculosis_H37Rv DDVGALVITGNGRVFSGGFDLKILTSGE-VQPAIDMLRGGFELAYRLLSY
MAV_4534|M.avium_104 DNAGALVITGNERVFSGGFDLKILTSGE-VQPAVNMLRGGFELAYRLLSY
Mvan_3990|M.vanbaalenii_PYR-1 DDVHVVVLTGAGRGFCSGADFGILSVLAESDSTFELMKHVSRPVQTLYHL
. ::: * . *..* *: : .
MUL_4319|M.ulcerans_Agy99 PGPTVAAINGAAIAGGCVLACACDRRLIGAEAAIGASEVLVGVAFPVVAL
MMAR_5243|M.marinum_M PGPTVAAINGAAIAGGCVLACACDRRLIGAEAAIGASEVLVGVAFPVVAL
Mflv_2036|M.gilvum_PYR-GCK RTPTIAAVAGYALGGGCELAMMCDILIAADTAKFGQPEIKLGVLPGMGGS
TH_4102|M.thermoresistible__bu RTPTIAAVAGYALGGGCELAMMCDLLIAADTAKFGQPEIKLGVLPGMGGS
MAB_1185c|M.abscessus_ATCC_199 RTPTIAAVSGYALGGGCELAMMCDVIIAAENATFGQPEIKLGVLPGMGGS
MLBr_02402|M.leprae_Br4923 RTPMIAAVAGYALGGGCELAMMCDLLIAADTAKFGQPEIKLGVLPGMGGS
MSMEG_2328|M.smegmatis_MC2_155 PKPTVAAVTGYALGSGMNLALAADWRVSGDNAKFGATEILAGLAPRGGGG
Mb0649c|M.bovis_AF2122/97 PKPVVMACTGHAIAMGAFLLSCGDHRVAAHAYNIQANEVAIGMTIPYAAL
Rv0632c|M.tuberculosis_H37Rv PKPVVMACTGHAIAMGAFLLSCGDHRVAAHAYNIQANEVAIGMTIPYAAL
MAV_4534|M.avium_104 PKPVVMACTGHAIAMGAFLLSSGDHRVAAHAYNIQANEVAIGMVIPYAAM
Mvan_3990|M.vanbaalenii_PYR-1 PQLTIAAVNGPAAGAGWGLAMACDIRVAAASARFGATFARMGLGPDYGLS
: * * * . * * * : . : *:
MUL_4319|M.ulcerans_Agy99 EVMRYACG-DRAEEVLLGGRSYRGIEAVSRGLAHEVVAED-LLGVATAQA
MMAR_5243|M.marinum_M EVMRYACG-DRAEEVLLGGRSYRGIEAVSRGLAHEVVAED-LLGVATAQA
Mflv_2036|M.gilvum_PYR-GCK QRLTRAIGKAKAMDLILTGRNMDAEEAERAGLVSRVVPADSLLSEAKAVA
TH_4102|M.thermoresistible__bu QRLTRAIGKAKAMDLILTGRTIDAEEAERSGLVSRVVPADKLLDEAKETA
MAB_1185c|M.abscessus_ATCC_199 QRLTRAIGKAKAMDMILTGRNMDAAEAERSGLVSRVVATESLLDEAKAVA
MLBr_02402|M.leprae_Br4923 QRLTRAIGKAKAMDLILTGRTIDAAEAERSGLVSRVVLADDLLPEAKAVA
MSMEG_2328|M.smegmatis_MC2_155 VRLADAIGTSKAKELVFSGRFVGAEEALEIGLVDEMVAPDHVYEAALAWA
Mb0649c|M.bovis_AF2122/97 EIMKLRLTRSAYQQATGLAKTFFGETALAAGFIDEIALPEVVVSRAEEAA
Rv0632c|M.tuberculosis_H37Rv EIMKLRLTRSAYQQATGLAKTFFGETALAAGFIDEIALPEVVVSRAEEAA
MAV_4534|M.avium_104 EIMKLRLTPSAYQQASGLAKTFFGETALAAGFVDEIVLPEVVVDRAEEAA
Mvan_3990|M.vanbaalenii_PYR-1 KTLPQAIGRDRALELLTTARIIDADEASAIGAVS--AVVDDVLAHTVRLA
: : .: . * * . : : : *
MUL_4319|M.ulcerans_Agy99 SELGDIPADAYRHTKAQLRAPARARMRDGG-DIDREVRQIWG-SAQTQQR
MMAR_5243|M.marinum_M SELGDIPADAYRHTKAQLRAPAQARMRDGG-DIDREVRQIWG-SAQTQQR
Mflv_2036|M.gilvum_PYR-GCK QTIAGMSLSASRMAKEAVDRAFETTLSEGLLYERRLFHSAFA-TDDQTEG
TH_4102|M.thermoresistible__bu ATIAAMSLSAARMAKEAVNRAFETSLAEGLLFERRVFHSAFA-TADQSEG
MAB_1185c|M.abscessus_ATCC_199 KTISEMSLSASMMAKEAVNRAFESSLAEGLLFERRIFHSAFG-TADQSEG
MLBr_02402|M.leprae_Br4923 TTISQMSRSATRMAKEAVNRSFESTLAEGLLHERRLFHSTFV-TDDQSEG
MSMEG_2328|M.smegmatis_MC2_155 RRFSDHPVDVLAAAKASLNGPVN---WRGPRPTDRMVDRP----------
Mb0649c|M.bovis_AF2122/97 REFAGLNQHAHAATKLRSRADALTAIRAGIDGIAAEFGL-----------
Rv0632c|M.tuberculosis_H37Rv REFAGLNQHAHAATKLRSRADALTAIRAGIDGIAAEFGL-----------
MAV_4534|M.avium_104 AEFAELNQRAHAATKLRARADALAAIRAGIDGIEAEFGL-----------
Mvan_3990|M.vanbaalenii_PYR-1 TEVARAPGRTVRSVKRTMRSAAAADFDTAVGVIEARAQAELFNHPDFMDV
.. . .* .
MUL_4319|M.ulcerans_Agy99 IAGYLERLRRR-----------
MMAR_5243|M.marinum_M IAGYLERLRRR-----------
Mflv_2036|M.gilvum_PYR-GCK MNAFSEKRSPNFTHR-------
TH_4102|M.thermoresistible__bu MAAFTEKRPPKFTHR-------
MAB_1185c|M.abscessus_ATCC_199 MAAFVEKRPANFIHR-------
MLBr_02402|M.leprae_Br4923 MAAFIEKRAPQFTHR-------
MSMEG_2328|M.smegmatis_MC2_155 ----------------------
Mb0649c|M.bovis_AF2122/97 ----------------------
Rv0632c|M.tuberculosis_H37Rv ----------------------
MAV_4534|M.avium_104 ----------------------
Mvan_3990|M.vanbaalenii_PYR-1 AAGWVAQVAGKAGPGHRPPVSP