For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
EKVPALYRVAAWVVIVAGITFIASTLFFAGALVFGHGACHHHRHHHHGMFKPGGSGGPGGPGGPGRMFFL PGPPPPGMGPGMGPGMGPGMSPGPHGGATGPVQTPSAPAPSTPPARPYRRAAR*
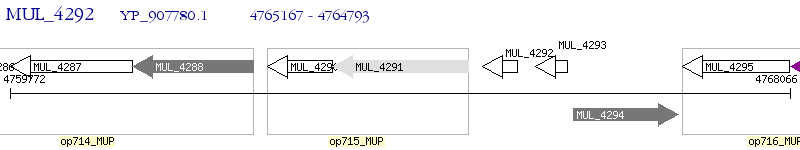
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_4292 | - | - | 100% (124) | hypothetical protein MUL_4292 |
| M. ulcerans Agy99 | MUL_4293 | - | 7e-14 | 36.13% (119) | hypothetical protein MUL_4293 |
| M. ulcerans Agy99 | MUL_2957 | - | 4e-07 | 43.08% (65) | PE-PGRS family protein |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb3732c | - | 1e-30 | 54.70% (117) | proline rich protein |
| M. gilvum PYR-GCK | Mflv_1307 | - | 2e-11 | 42.73% (110) | hypothetical protein Mflv_1307 |
| M. tuberculosis H37Rv | Rv3706c | - | 2e-13 | 36.61% (112) | proline rich protein |
| M. leprae Br4923 | - | - | - | - | - |
| M. abscessus ATCC 19977 | MAB_2580c | - | 6e-07 | 54.55% (44) | hypothetical protein MAB_2580c |
| M. marinum M | MMAR_5218 | - | 6e-70 | 96.69% (121) | hypothetical protein MMAR_5218 |
| M. avium 104 | MAV_0397 | - | 2e-27 | 46.15% (143) | hypothetical protein MAV_0397 |
| M. smegmatis MC2 155 | MSMEG_6254 | - | 2e-13 | 40.17% (117) | hypothetical protein MSMEG_6254 |
| M. thermoresistible (build 8) | TH_3578 | - | 5e-10 | 33.63% (113) | PUTATIVE - |
| M. vanbaalenii PYR-1 | Mvan_5408 | - | 8e-16 | 46.51% (129) | hypothetical protein Mvan_5408 |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_4292|M.ulcerans_Agy99 --------------------------------------------MEKV--
MMAR_5218|M.marinum_M MSET----SEPATPPPAVAT----------------AAPPPPPPVEKV--
MAV_0397|M.avium_104 MSQTPEPQAEPTTPEPTPAA----------------AVPPPPPPPERVKP
MSMEG_6254|M.smegmatis_MC2_155 MSETPSDHPTTPVGTPAETPTATSGPVAPPPVAPPPAAVPVAPAEAPRRH
TH_3578|M.thermoresistible__bu MTDAPEPRPTEPQAPPAQ-----------------PAAAPPAVSNAPR-S
Rv3706c|M.tuberculosis_H37Rv MRHMSETS-ETPTPPPHQ-------------------------------T
Mflv_1307|M.gilvum_PYR-GCK MSDVTSHQ-SDPATTPVP----------------APPSATAPPVVVPQES
Mb3732c|M.bovis_AF2122/97 MTETPQPAAPPPSAATTSP----------------------PPSPQQEKP
Mvan_5408|M.vanbaalenii_PYR-1 MTETPESRTEPTAVATEQGR--------------------GELSPGSDRP
MAB_2580c|M.abscessus_ATCC_199 --------------------------------------------------
MUL_4292|M.ulcerans_Agy99 PALYRVAAWVVIVAGITFIASTLFFAGALVFGHGACHHHR----------
MMAR_5218|M.marinum_M PALYRVAAWVVIVAGITFIASTLFFAGALVFGHGACHHHR----------
MAV_0397|M.avium_104 PRLYTAAAWVVIVAGIVFIISSVFFVGAFIWKH--SYYHC----------
MSMEG_6254|M.smegmatis_MC2_155 SRVVQAAAWVGIVAGVVFIVAVVFGTGFVLGKNSG---------------
TH_3578|M.thermoresistible__bu SRLPQVAAWIAIVAGVLVIMAVIFFAGFFLGRVSDG--------------
Rv3706c|M.tuberculosis_H37Rv PKVFKAAAWVAIAAGTVFIVAVIFFTGYILGKHAG---------------
Mflv_1307|M.gilvum_PYR-GCK NRLNKAAAWVGIAAGAVFIVAVVFFAGFLLGQNVDG--------------
Mb3732c|M.bovis_AF2122/97 PRLYRAAAWVVIVAGIVFTVAVIFFSGALVLGQGKCPYHR----YYHHGM
Mvan_5408|M.vanbaalenii_PYR-1 NRLTQSAAWVGIVAGVVFIVAVIFFSGFFVGKQYGGGYRDGRGMSYPGGM
MAB_2580c|M.abscessus_ATCC_199 MKMARALTYGAVAGG--LTLGILGFGASVASAEP----------------
: :: :..* . . : . .
MUL_4292|M.ulcerans_Agy99 HHHHGMFKPGGSG-----------GPGG---PGGPGR---MFFLPGPPPP
MMAR_5218|M.marinum_M HHHHGMFKPGGSG-----------GPGG---PGGPGGPGGMFFLPGPPPP
MAV_0397|M.avium_104 HHHHGVFHPGGPGPGNGPWQYGGPGPGNGPWPGGPGGSGAWYGGPGGPDN
MSMEG_6254|M.smegmatis_MC2_155 ---PGHHHRGHDRPE------------------------IMMFHRGGPPP
TH_3578|M.thermoresistible__bu --FGGHHHR-------------------------------MHWHDRMPSR
Rv3706c|M.tuberculosis_H37Rv --HGGFHHR---------------------------------QHHQHPAM
Mflv_1307|M.gilvum_PYR-GCK --GGRAHHGG------------------------------PGMMQPGPAG
Mb3732c|M.bovis_AF2122/97 FRPVGPVAPGPGMG--------------------------WVFG--FPG-
Mvan_5408|M.vanbaalenii_PYR-1 MGPGGMMGPGGMMGP------------------------GGMMGPGGPGG
MAB_2580c|M.abscessus_ATCC_199 --PPPIPVPG-------------------------------LPGPGGPP-
*
MUL_4292|M.ulcerans_Agy99 G---MGPGMGPG----MGPGMSPGPHG-----GATGPVQ-TPSAPAPSTP
MMAR_5218|M.marinum_M G---MGPGMGPG----MGPGMSPGPHG-----GATGPVQ-TPSAPAPSTP
MAV_0397|M.avium_104 GPWPGGPGLIVP----MPPGGPGGPGGPVSGPGVSGPGQPSPVSPAPSTP
MSMEG_6254|M.smegmatis_MC2_155 MMPGMGPGHMRPGGGMFGPGGAPAERGE--FRGPQAPD-RSETPT-TAPA
TH_3578|M.thermoresistible__bu YFDHPGPGPR------FSP-SAPGTR-------PAPPT-EPAAPSPTATP
Rv3706c|M.tuberculosis_H37Rv MLR---PG---------SPHGGPAAVRP--GPGPGGPGQVPSSVSPPATP
Mflv_1307|M.gilvum_PYR-GCK FPMGPPGGFHHG-----PGFAGPSGGGP--GMDAPRPQSPTTGNAPTPSP
Mb3732c|M.bovis_AF2122/97 --GPPPPG--MGPG---FPG---GPG----GP-AVGP-TGPGPTTAPARP
Mvan_5408|M.vanbaalenii_PYR-1 MMGPGGPGGMMGPGS-WGPGGMMGPGSW--GPGMMGPGQQPSPTTTPAVP
MAB_2580c|M.abscessus_ATCC_199 -----GPG--------IPGPGVPGPG-----PGIPGPGVPGPDGPGPDGP
* . * . . .
MUL_4292|M.ulcerans_Agy99 PARPYRRAAR
MMAR_5218|M.marinum_M PARP------
MAV_0397|M.avium_104 APRP------
MSMEG_6254|M.smegmatis_MC2_155 RP--------
TH_3578|M.thermoresistible__bu RP--------
Rv3706c|M.tuberculosis_H37Rv AP--------
Mflv_1307|M.gilvum_PYR-GCK RP--------
Mb3732c|M.bovis_AF2122/97 ----------
Mvan_5408|M.vanbaalenii_PYR-1 TTPQR-----
MAB_2580c|M.abscessus_ATCC_199 FFH-------