For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MVTDAPLKFGVFITPFHPTGQSPTVALEYDMERVVALDRLGYDEAWFGEHHSGGYELIACPEVFIAAAAE RTKHIRLGTGVVSLPYHHPLMVADRWVLLDHLTRGRVVFGTGPGALPSDAYMMGIDPIDQRRMMQESLEA ILALLRAAPGERIDRHSDWFTLRDAQLHIRPYTWPYPEISTAAMISPSGPRLAGSLGTSLLSLSMSVPGG YAALENTWQVLCEQAAKVGRAEPNRRDWRVLSIMHLSDTPRPGDPRLHLRAAGFLEVLRRDFGAAGFVPL SNATDGAEGTREFAEDYAAKGNCCIGTPEDEIAYIEDLLNRSGGFGTLLLLGHDWASPQATFHSYDLFAR KVIPYFKGQLEAPRASHEWARDKREDLIGRAGQAVVAAITEHVARFSSLRSPVTPTSRHVGLAGGRRARH HPGMKPISGRGRRGGEAARGCAMSRQAAKDMLAGVGGRGGSGNGAERNGCNAAQPGHRTRPSVGYRQTPD LGGPANPPETTRHARQRQFRQRLWQGLDRLGGFLARVRGSREETVGQTVDVFQRRGGEPEGRNQPDIGVR
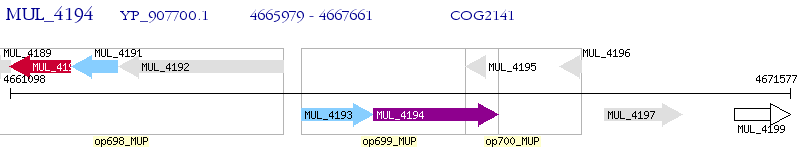
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_4194 | - | - | 100% (560) | monooxygenase |
| M. ulcerans Agy99 | MUL_2949 | - | 7e-76 | 42.15% (382) | monooxygenase |
| M. ulcerans Agy99 | MUL_2890 | - | 6e-38 | 30.19% (371) | oxygenase |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb1971 | - | 2e-37 | 29.97% (377) | monooxygenase |
| M. gilvum PYR-GCK | Mflv_4429 | - | 2e-95 | 46.72% (396) | luciferase family protein |
| M. tuberculosis H37Rv | Rv3618 | - | 0.0 | 83.85% (390) | monooxygenase |
| M. leprae Br4923 | MLBr_00131 | - | 1e-08 | 32.06% (131) | putative oxidoreductase |
| M. abscessus ATCC 19977 | MAB_0527c | - | 1e-163 | 70.45% (396) | monooxygenase |
| M. marinum M | MMAR_5115 | - | 0.0 | 94.13% (392) | monooxygenase |
| M. avium 104 | MAV_0538 | - | 0.0 | 80.21% (389) | limonene 1,2-monooxygenase |
| M. smegmatis MC2 155 | MSMEG_6107 | - | 1e-179 | 77.78% (387) | limonene 1,2-monooxygenase |
| M. thermoresistible (build 8) | TH_0993 | - | 1e-179 | 77.12% (389) | POSSIBLE MONOOXYGENASE |
| M. vanbaalenii PYR-1 | Mvan_1925 | - | 6e-97 | 47.63% (401) | luciferase family protein |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_4194|M.ulcerans_Agy99 MVTDAPLKFGVFITPFHPTGQSPTVALEYDMERVVALDRLGYDEAWFGEH
MMAR_5115|M.marinum_M -MTDAPLKFGVFITPFHPTGQSPTVALEYDMERVVALDRLGYDEAWFGEH
Rv3618|M.tuberculosis_H37Rv --MKAPLRFGVFITPFHPTGQSPTVALQYDMERVVALDRLGYDEAWFGEH
MAV_0538|M.avium_104 --MSAPLRFGVFITPFHPVGQSPTVALEYDLERVVALDRLGFDEAWFGEH
MSMEG_6107|M.smegmatis_MC2_155 ---MRPLNFGVFITPFHPAGQSPTTALQYDMDRVEALDRLGYDEAWFGEH
TH_0993|M.thermoresistible__bu --VARPLNFGVFITPFHPVGQSPTLALEYDLERTVALDRLGYDEVWFGEH
MAB_0527c|M.abscessus_ATCC_199 -MSAPPLRFGAFITPFHPVGQNPTTALEYDLDRVVALDRLGYHEAWFGEH
Mflv_4429|M.gilvum_PYR-GCK ------MRFGNFMAPFHPVGQNPTLALERDLDLIVAMDRLGFHEAWVGEH
Mvan_1925|M.vanbaalenii_PYR-1 ------MRFGNFMAPFHPVGQNPTLALERDLDLIAAMDRLGFHEAWVGEH
Mb1971|M.bovis_AF2122/97 ------MEIGIFLMPAHPPERTLYDATRWDLDVIELADQLGYVEAWVGEH
MLBr_00131|M.leprae_Br4923 ---MAGFRFGFVDALVHTLFPPSLPARAS----IVSGAVLGADSYWVGDH
:.:* . *. * ** . *.*:*
MUL_4194|M.ulcerans_Agy99 HSGGYELIACPEVFIAAAAERT----------------------KHIRLG
MMAR_5115|M.marinum_M HSGGYELIACPEVFIAAAAERT----------------------KHIRLG
Rv3618|M.tuberculosis_H37Rv HSGGYELIACPEVFIAAAAERT----------------------THIRLG
MAV_0538|M.avium_104 HSGGYELIACPEVFIATAAERT----------------------KHIRLG
MSMEG_6107|M.smegmatis_MC2_155 HSGGYELIACPEVFIAAAAERT----------------------RHIRLG
TH_0993|M.thermoresistible__bu HSGGYELIASPEVFIAAAAERT----------------------RHVRLG
MAB_0527c|M.abscessus_ATCC_199 HSGGYELISCPEVFIATAAERT----------------------KHIKLG
Mflv_4429|M.gilvum_PYR-GCK HSAGFEIIASPEVFLGVAAERT----------------------KHIRLG
Mvan_1925|M.vanbaalenii_PYR-1 HSAGFEIIASPEVFIGIAAERT----------------------KHIKLG
Mb1971|M.bovis_AF2122/97 FTVPWEPICAPDLLLAQALLRT----------------------QQIKLA
MLBr_00131|M.leprae_Br4923 LNALVPRSVATPKYLGVAAKVVPKIDANYEPWTMLGNLAAGNRLNGLRLG
. .. :. * . ::*.
MUL_4194|M.ulcerans_Agy99 TGVVSLPYHHPLMVADRWVLLDHLTRGRVVFGTGPGALPSDAYMMGIDPI
MMAR_5115|M.marinum_M TGVVSLPYHHPLMVADRWVLLDHLTRGRVMFGTGPGALPSDAYMMGIDPI
Rv3618|M.tuberculosis_H37Rv TGVVSLPYHHPLMVADRWVLLDHLTRGRVMFGTGPGALPSDAYMMGIDPV
MAV_0538|M.avium_104 TGVVSLPYHHPLMVADRWVLLDHLTRGRVMFGTGPGALPSDAYMMGIDPV
MSMEG_6107|M.smegmatis_MC2_155 TGVVSLPYHHPLMVADRWVLLDHLTRGRVMFGTGPGALPSDAYMMGIDPV
TH_0993|M.thermoresistible__bu TGVVSLPYHHPLMVADRWVLLDHITRGRVMFGAGPGALPSDAYMMGIDPV
MAB_0527c|M.abscessus_ATCC_199 TGVVSLPYHHPLMVVDRWILLDHITRGRVIFGTGPGALPTDAYMMGIDPV
Mflv_4429|M.gilvum_PYR-GCK TGVSSLPYHHPLMLADRMVLLDHLTRGRVMLGCGPGQLTSDAHMLGIPAD
Mvan_1925|M.vanbaalenii_PYR-1 TGVSSLPYHHPLLLADRMVLLDHLTRGRVMLGCGPGQLTSDAHMLGIPAD
Mb1971|M.bovis_AF2122/97 PGAHLLPYHHPVELAHRVAYFDHLAQGRFMLGVGASGIPGDWALYDVDGK
MLBr_00131|M.leprae_Br4923 VCVTDAGRRNPAVTAQAAATLHLLTRGKAMLGIGVGEREGNE-PYGVEWT
. ::* .. :. :::*: ::* * . : .:
MUL_4194|M.ulcerans_Agy99 --DQRRMMQESLEAILALLRAAPGERIDRHSDWFTLR--------DAQLH
MMAR_5115|M.marinum_M --DQRRMMQESLEAILALLRAAPGERIDRHSDWFTLR--------DAQLH
Rv3618|M.tuberculosis_H37Rv --EQRRMMQESLEAILALFRAAPDERIDRHSDWFTLR--------EAQLH
MAV_0538|M.avium_104 --DQRHMMQESLEAILALFRAAPTERIDRHSDWFTLR--------DAQLH
MSMEG_6107|M.smegmatis_MC2_155 --DQRPMMQESLEAILALFRAEPGERIDRKTDWFTLR--------DAALH
TH_0993|M.thermoresistible__bu --NQRPMMQEALEAILALLRAAPDERISRETDWFTLR--------DAQLH
MAB_0527c|M.abscessus_ATCC_199 --DQRRMQEESLEAILALLRATPQERINRETSWFTLR--------DAQLQ
Mflv_4429|M.gilvum_PYR-GCK --EQRPRMEQCLDAIVRLLRG---ETVTMHTDGFTLQ--------DARLQ
Mvan_1925|M.vanbaalenii_PYR-1 --AQRPRMEQSLDAIVRLLRG---ETVTMHTDGFTLQ--------DARLQ
Mb1971|M.bovis_AF2122/97 NGEHREMTREALEIMLRIWTE--DEPWEHRGKYWNANGIAPMFEGLMRRH
MLBr_00131|M.leprae_Br4923 --KPVARFQEALATIRALWDSN-GELVSRESQFFPLH--------NALFD
.:.* : : * . . : . .
MUL_4194|M.ulcerans_Agy99 IRPYTWPYPEISTAAMIS-PSGPRLAGSLGTSLLSLSMSVPGGYAALENT
MMAR_5115|M.marinum_M IRPYTWPYPEISTAAMIS-PSGPRLAGSLGTSLLSLSMSVPGGYAALENT
Rv3618|M.tuberculosis_H37Rv IRPYTWPYPEIATAAMIS-PSGPRLAGALGTSLLSLSMSVPGGYAALETA
MAV_0538|M.avium_104 IRPYTWPYPEISTAAMIS-PSGPRLAGALGTSLLSLSMSVPGGYAALETT
MSMEG_6107|M.smegmatis_MC2_155 IRPYTWPYPEISTAAMIS-PSGPRLAGKLGTSLLSLSMSVPGGYAAVETT
TH_0993|M.thermoresistible__bu IRPYTWPYPEISTAAMIS-PSGPRLAGSLGTSLLSLSMSVPGGYAAVENT
MAB_0527c|M.abscessus_ATCC_199 LRPYTHPYPEILTAAMFS-PSGPRLAGKLGAGLLSLSASIPGGFAALENA
Mflv_4429|M.gilvum_PYR-GCK LKPYSEPCFDIAVAASFS-PTGPRGAGRHGIGMLSIAATARQGMDLLAQH
Mvan_1925|M.vanbaalenii_PYR-1 LKPYSDPCFDIAVAASFS-PTGPRGAGRHGIGMLSIAATAKQGMDLLAQH
Mb1971|M.bovis_AF2122/97 IKPYQKPHPPIGVTGFSAGSETLKLAGERGYIPMSLDLNT----EYVATH
MLBr_00131|M.leprae_Br4923 LPPYRGKWPEIWVAAHGP--RMLQATGRYADAWIPIVLVRP---TDYSCA
: ** * .:. . : :* . :.:
MUL_4194|M.ulcerans_Agy99 WQVLCEQAAKVGRAEPNRRDWRVLSIMHLSDTPRPGDPRLHLRAAG--FL
MMAR_5115|M.marinum_M WQVVCEQAAKVGRAEPNRRDWRVLSIMHLSDTR---DQAIQDCIYG--LP
Rv3618|M.tuberculosis_H37Rv WGVVREQAAKAGRGEPDRADWRVLSIMHLSDSR---DQAIDDCTYG--LP
MAV_0538|M.avium_104 WDVVREQAAKAGRDEPDRGDWRVLSIMHIADSR---EQAIDDCTYG--LQ
MSMEG_6107|M.smegmatis_MC2_155 WNVVRDEAAAAGRPEPDRENWRVLGIVHLADTR---EQAIEDCTYG--LQ
TH_0993|M.thermoresistible__bu WDVVVQQAAKAGRPEPDRANWRVLGIVHLADTR---EQALQDCTYG--LQ
MAB_0527c|M.abscessus_ATCC_199 WEVVREQAEIHGHPEPDRSSWRVLGIMHLAETR---DQAIADCTYG--LP
Mflv_4429|M.gilvum_PYR-GCK WNTWDEVAREHG-HQADRSKWRLVGPMHLAETR---EQAERDVEYG--IA
Mvan_1925|M.vanbaalenii_PYR-1 WNTWEEVAREHG-HVADRAKWRLVGPMHLAETR---EQAERDVEYG--IA
Mb1971|M.bovis_AF2122/97 WDAVEEGALRSG-RTPDRRDWRLVREVLVAETDEQAFRYAVDGTMGRAMR
MLBr_00131|M.leprae_Br4923 LEVVRTAASDAG-RDPMSITPAAVRGIITGRTRDDVDEALDSVLVRMIAL
. * * . : : . :
MUL_4194|M.ulcerans_Agy99 EVLRRDFGAAGFVPLSNATDGAE---GTREFAEDYAAKGNCCIGTPEDEI
MMAR_5115|M.marinum_M DFS-KYFGAAGFVPLSNATDGAE---GTREFVEDYAAKGNCCIGTPEDAI
Rv3618|M.tuberculosis_H37Rv DFS-RYFGAAGFVPLANTVEGTQ---SSREFVEQYAAKGNCCIGTPDDAI
MAV_0538|M.avium_104 DFA-NYFGAAGFVPLANSVEGTQ---TPHEFVAEYAAKGNCCIGTPEDAI
MSMEG_6107|M.smegmatis_MC2_155 DFA-NYFGAAGFVPLSNQAEGGG---TPYEFVENYASKGNCCIGTPDDAI
TH_0993|M.thermoresistible__bu DFA-NYFGAAGFVPLANEVAGGQ---SPREFVEAYAEKGNCCIGTPGDAI
MAB_0527c|M.abscessus_ATCC_199 DYA-HYFGAVGVLPLANSADGADGAIDPHEFVKAYADEGNCCIGTPEDAI
Mflv_4429|M.gilvum_PYR-GCK EFS-RYFT---HILPSGPVQGDT----PEEIIANNRQSGFAVIGTPDDAI
Mvan_1925|M.vanbaalenii_PYR-1 QFS-QYFS---HILPAGPVQGDT----PAEIIANNHASGFAVIGTPQDAI
Mb1971|M.bovis_AF2122/97 EYVLPTFRMFGMTKFYKHNPSVP----DDEVTPEYLAENTFVVGSVQTVV
MLBr_00131|M.leprae_Br4923 GVPGEAWARHGVEHPMGADFAGV-----QDIIPQTIDEETVVSYAAKVPA
: . :. . :
MUL_4194|M.ulcerans_Agy99 AYIEDLLNRS-GGFGTLLLLGHDWAS-PQATFHSYDLFARKVIPYFKGQL
MMAR_5115|M.marinum_M AYIEDLLNRS-GGFGTLLLLGHDWAS-PQATFHSYDLFARKVIPYFKGQL
Rv3618|M.tuberculosis_H37Rv AHIEDLLHRS-GGFGTLLLLGHDWAP-PPATFHSYELFARAVIPYFKGQL
MAV_0538|M.avium_104 AHIEDLLDRS-GGFGTLLMLGHDWAS-PEATYHCYDLMARKVIPHFKRQL
MSMEG_6107|M.smegmatis_MC2_155 VYIQDLLDRS-GGFGTFLLLGHDWAP-PAATFHSYELFAREVIPHFKGQL
TH_0993|M.thermoresistible__bu VYLSDLLERS-GGFGTFLMLGHDWAS-PQATYHSYELFAREVIPHFKGQL
MAB_0527c|M.abscessus_ATCC_199 EYIQGLLDTS-GGFGTFLMLGHDWAA-PEATFKSYELFARKVIPHFTGQL
Mflv_4429|M.gilvum_PYR-GCK AKISELVEASDGGFGAFLLFDHDWAP-PAAKLRSYELFAQYVIPHFTGQL
Mvan_1925|M.vanbaalenii_PYR-1 AKIEELVEASNGGFGTFLLFDHDWAP-PAAKLRSYELFAEYVIPHFTGQL
Mb1971|M.bovis_AF2122/97 DKLEATYDQV-GGFGHLLILGFDYSDNPGPWKESLRLLAHEVMPRLNARL
MLBr_00131|M.leprae_Br4923 ALMKEVLFSG--TPEEVIDQVAEWRD--HGLKYLVVINGSLVNSSLRKTV
:. .: :: : . * . : :
MUL_4194|M.ulcerans_Agy99 EAPRASHEWARDKREDLIGRAGQAVVAAITEHVARFSSLRSPVTPTSRHV
MMAR_5115|M.marinum_M EAPRASHEWARDKREDLIGRAGQAVVAAITEHVTER--------------
Rv3618|M.tuberculosis_H37Rv AAPRASHEWARGKRDQLIGRAGEAVVKAITEHVAEQ--------------
MAV_0538|M.avium_104 DASRASHDWAKGMRDQLLGRAGEAVVKAISEHVDEH--------------
MSMEG_6107|M.smegmatis_MC2_155 TAARASHDWAKGMRDQLLGRAGQAIVNAITEHTQAH--------------
TH_0993|M.thermoresistible__bu TAPRASHDWAKQMRGQLLGRAGEAIVKAIGEHTSEQ--------------
MAB_0527c|M.abscessus_ATCC_199 TAAQASHDWAQAKRGELLGRAGDAVAKAIGEHVAQK--------------
Mflv_4429|M.gilvum_PYR-GCK SAPIASRDWVTESGRVFVDQATHAIGRAIESHAAEQ--------------
Mvan_1925|M.vanbaalenii_PYR-1 SAPIASRDWVTESGRTFVDQAAHAIGKAIEDHAAEQ--------------
Mb1971|M.bovis_AF2122/97 ATKPATAVV-----------------------------------------
MLBr_00131|M.leprae_Br4923 SALLPHARVLRGLKKL----------------------------------
: .
MUL_4194|M.ulcerans_Agy99 GLAGGRRARHHPGMKPISGRGRRGGEAARGCAMSRQAAKDMLAGVGGRGG
MMAR_5115|M.marinum_M ------------------GEGQT---------------------------
Rv3618|M.tuberculosis_H37Rv ------------------GEAGS---------------------------
MAV_0538|M.avium_104 ------------------KDAVH---------------------------
MSMEG_6107|M.smegmatis_MC2_155 -------------------PAENEN-------------------------
TH_0993|M.thermoresistible__bu -------------------EQR----------------------------
MAB_0527c|M.abscessus_ATCC_199 --------------------ASH---------------------------
Mflv_4429|M.gilvum_PYR-GCK -------------------QAKQSRTSD----------------------
Mvan_1925|M.vanbaalenii_PYR-1 -------------------QAKSPTPS-----------------------
Mb1971|M.bovis_AF2122/97 --------------------------------------------------
MLBr_00131|M.leprae_Br4923 --------------------------------------------------
MUL_4194|M.ulcerans_Agy99 SGNGAERNGCNAAQPGHRTRPSVGYRQTPDLGGPANPPETTRHARQRQFR
MMAR_5115|M.marinum_M --------------------------------------------------
Rv3618|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0538|M.avium_104 --------------------------------------------------
MSMEG_6107|M.smegmatis_MC2_155 --------------------------------------------------
TH_0993|M.thermoresistible__bu --------------------------------------------------
MAB_0527c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_4429|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1925|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb1971|M.bovis_AF2122/97 --------------------------------------------------
MLBr_00131|M.leprae_Br4923 --------------------------------------------------
MUL_4194|M.ulcerans_Agy99 QRLWQGLDRLGGFLARVRGSREETVGQTVDVFQRRGGEPEGRNQPDIGVR
MMAR_5115|M.marinum_M --------------------------------------------------
Rv3618|M.tuberculosis_H37Rv --------------------------------------------------
MAV_0538|M.avium_104 --------------------------------------------------
MSMEG_6107|M.smegmatis_MC2_155 --------------------------------------------------
TH_0993|M.thermoresistible__bu --------------------------------------------------
MAB_0527c|M.abscessus_ATCC_199 --------------------------------------------------
Mflv_4429|M.gilvum_PYR-GCK --------------------------------------------------
Mvan_1925|M.vanbaalenii_PYR-1 --------------------------------------------------
Mb1971|M.bovis_AF2122/97 --------------------------------------------------
MLBr_00131|M.leprae_Br4923 --------------------------------------------------