For questions or suggestions e-mail us at: ioerger@cs.tamu.edu
MSIVAVAIFVVAYALIASERVNKTLVALAGAAIVVMLPVINSEDVFYSHETGVDWDVIFLLLGMMIIVSV LRQTGVFEYVAICAAKRARGKPLRIMILLVIVTAVASALLDNVTTVLLIAPVTLLVCDRLVINPAPYLMA EAFASNIGGTATLVGDPPNIVIASKGGLTFNDFLIHLAPIVVIVMAVFILMLPRLFPGSFEADPERVADV MSLEEKEAIRDHGLLIKCGIVLLLVFTAFVANSALHVKPSMVALIGAGILVVVSRLDHSDYLSSVEWDTL LFFVGLFIMVGALVKTGVVTQVAQMAVTATGGNTLLATMVILASSLVISGIVNNVPYAATMTPIVAELVP VLVDKGNPDVLWWALALGTDFGGDLTAIGASANVIVLGIAKRADNPISFWEFTRKGIVVTTMSLVLAAIY LWVRYFVLS
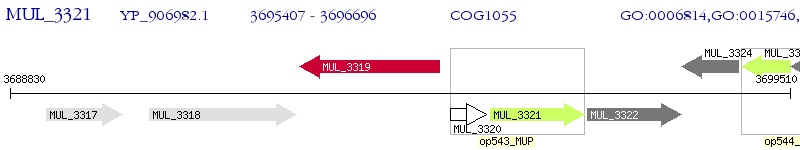
Operon Prediction Model: Genebank
Paralogs
| species | id | gene | e-value | identity (len) | annotation |
| M. ulcerans Agy99 | MUL_3321 | arsA | - | 100% (429) | arsenic-transport integral membrane protein ArsA |
| M. ulcerans Agy99 | MUL_4154 | arsB2 | 9e-25 | 24.58% (419) | arsenical pump integral membrane protein ArsB2 |
Closest Orthologs (e-value cutoff: 1e-4)
| species | id | gene | e-value | identity (len) | annotation |
| M. bovis AF2122 / 97 | Mb2703 | arsA | 1e-175 | 71.73% (428) | arsenic-transport integral membrane protein ArsA |
| M. gilvum PYR-GCK | Mflv_1448 | - | 5e-31 | 26.06% (426) | arsenical pump membrane protein |
| M. tuberculosis H37Rv | Rv2684 | arsA | 1e-175 | 71.73% (428) | arsenic-transport integral membrane protein ArsA |
| M. leprae Br4923 | MLBr_01036 | - | 1e-177 | 70.40% (429) | putative membrane transport protein |
| M. abscessus ATCC 19977 | - | - | - | - | - |
| M. marinum M | MMAR_2030 | arsA | 0.0 | 99.30% (429) | arsenic-transport integral membrane protein ArsA |
| M. avium 104 | MAV_3579 | - | 0.0 | 77.99% (427) | citrate transporter |
| M. smegmatis MC2 155 | MSMEG_0851 | - | 1e-130 | 51.89% (424) | citrate transporter |
| M. thermoresistible (build 8) | TH_3461 | - | 2e-28 | 25.53% (423) | PUTATIVE putative Arsenical pump membrane protein |
| M. vanbaalenii PYR-1 | Mvan_5336 | - | 4e-33 | 25.30% (423) | arsenical pump membrane protein |
CLUSTAL 2.0.9 multiple sequence alignment
MUL_3321|M.ulcerans_Agy99 ---MSIVAVAIFVVAYALIASERVNKTLVALAGAAIVVMLPVINSEDVFY
MMAR_2030|M.marinum_M ---MSIVAVAIFVVAYALIASERVNKTLVALAGAAIVVMLPVINSEDVFY
MAV_3579|M.avium_104 ---MSVIAIAVFVVAYALIASDRVNKTFVALAGAAVVITLPLIRSDDVFY
Mb2703|M.bovis_AF2122/97 ---MSVVAVTIFVAAYVLIASDRVNKTMVALTGAAAVVVLPVITSHDIFY
Rv2684|M.tuberculosis_H37Rv ---MSVVAVTIFVAAYVLIASDRVNKTMVALTGAAAVVVLPVITSHDIFY
MLBr_01036|M.leprae_Br4923 ---MSVIAITVFIVTYALIASDRVNKTLAALTGAAIVVTLPIINSEDVFY
MSMEG_0851|M.smegmatis_MC2_155 -MLAPVLALTIFVVAFWFIATERADKVKTVLVAAGLMTLLGLVPGAEVFY
Mflv_1448|M.gilvum_PYR-GCK MALTLSVAALAAVLACALLRPR--LQAIVAGLAAALVIVAGATTWRAAVG
Mvan_5336|M.vanbaalenii_PYR-1 MELILSVAALASVLAFALLRPHRWPEAVIAVPAAALVIASGAISWDDALA
TH_3461|M.thermoresistible__bu MAVTVASAALIAVLVSALTRPRGWPEAVVAVPAAALVVLTGAIALPDALA
* : . : . :. . .*. : .
MUL_3321|M.ulcerans_Agy99 SHETGVDWDVIFLLLGMMIIVSVLRQTGVFEYVAICAAKRARGKPLRIMI
MMAR_2030|M.marinum_M SHETGVDWDVIFLLLGMMIIVSVLRQTGVFEYVAIWAAKRARGKPLRIMI
MAV_3579|M.avium_104 SRETGIDWDVIFLLLGMMIIVSVLRQTGVFEYVAIWSAKRARGSPLRVMI
Mb2703|M.bovis_AF2122/97 SHDTGIDWDVIFLLVGMMIIVGVLRQTGVFEYTAIWAAKRARGSPLRIMI
Rv2684|M.tuberculosis_H37Rv SHDTGIDWDVIFLLVGMMIIVGVLRQTGVFEYTAIWAAKRARGSPLRIMI
MLBr_01036|M.leprae_Br4923 SHETGIDWEVIFLLLSMMIIVSVLRQTGVFEYVAIWTAKRSHGSPLRILL
MSMEG_0851|M.smegmatis_MC2_155 SEHEGIDWNVIFLLLGMMVIVGVVKQTGLFDFLAIWAAKRSRGKPFRLMV
Mflv_1448|M.gilvum_PYR-GCK EISE--LAPVVAFLAAVLVLARLCDDEGLFRAAGTLMARATSGGQSRLLT
Mvan_5336|M.vanbaalenii_PYR-1 EVGR--LAPVVGFLAAVLVLARLCDDVGLFHTAGVMMARATSGGQIRLLA
TH_3461|M.thermoresistible__bu VADR--LLPVVGFLAAVLVLARLCDDEGLFRAAGAAMARVSSGNPVRLLA
*: :* .::::. : : *:* . *: : * *::
MUL_3321|M.ulcerans_Agy99 LLVIVTAVASALLDNVTTVLLIAPVTLLVCDRLVINPAPYLMAEAFASNI
MMAR_2030|M.marinum_M LLVIVTAVASALLDNVTTVLLIAPVTLLVCDRLVINPAPYLMAEAFASNI
MAV_3579|M.avium_104 LLVLVTALASALLDNVTTVLLIAPVTLLVCDRLAIAAAPFLMAEVFASNV
Mb2703|M.bovis_AF2122/97 LLVLVSALASALLDNVTTVLLIAPVTLLVCDRLNINTTSFLMAEVFASNI
Rv2684|M.tuberculosis_H37Rv LLVLVSALASALLDNVTTVLLIAPVTLLVCDRLNINTTSFLMAEVFASNI
MLBr_01036|M.leprae_Br4923 LLVLVMALGSALLDNVTTVLLIAPVTLLVCERLTINAAPFLMAEVFASNI
MSMEG_0851|M.smegmatis_MC2_155 MLMIITAFASPVLDNVTIILLVAPVTVVICDRLRIPPQPYLIAEVLASNI
Mflv_1448|M.gilvum_PYR-GCK TVFAISAAVTAILSLDATVILLTPVVLATARALAVPARPHAYATAHLANS
Mvan_5336|M.vanbaalenii_PYR-1 SVFAIAAGVTAILSLDATVVLLTPVVLATARTLAVPPRPHAYATAHLANS
TH_3461|M.thermoresistible__bu MVFVVAAATTAILSLDATAVLLTPVVLATARRLGVAPRPHAYATAHLANS
:. : * :.:*. : :*::**.: . * : . .. * . :*
MUL_3321|M.ulcerans_Agy99 GGTATLVGDPPNIVIASKGGLTFNDFLIHLAPIVVIVMAVFILMLPRLFP
MMAR_2030|M.marinum_M GGTATLVGDPPNIVIASKGGLTFNDFLIHLAPIVVIVMAVFILMLPRLFP
MAV_3579|M.avium_104 GGAATLVGDPPNIIIASRGGLSFNDFLVHLAPIVVIVVGVLIALLPRLFP
Mb2703|M.bovis_AF2122/97 GGAATLVGDPPNIIVASRAGLTFNDFMLHLTPLVVIVLIALIAVLPRLF-
Rv2684|M.tuberculosis_H37Rv GGAATLVGDPPNIIVASRAGLTFNDFMLHLTPLVVIVLIALIAVLPRLF-
MLBr_01036|M.leprae_Br4923 GGAATLVGDPPNIIIASRAGFSFNDFLIHLTPIVIIVTVVLSALLPRLFR
MSMEG_0851|M.smegmatis_MC2_155 GGAATLIGDPPNIIIGSRAGLTFNDFLIHMAPIVIVIFALFVVFTRVLFR
Mflv_1448|M.gilvum_PYR-GCK ASLLLPVSNLTNLLAFSAAGLSFLHFAAAMTLPWIAAIVVEFLLLRWLFA
Mvan_5336|M.vanbaalenii_PYR-1 ASLLLPVSNLTNLLAFGAAGLSFLHFAAAMSLPWLAAIAVEFVLLRWFFA
TH_3461|M.thermoresistible__bu ASLLLPVSNLTNLLAFGVCGLSFTRFTALMTLPWLAAIACEFVLLRRLFR
.. :.: .*:: . *::* * :: : . :*
MUL_3321|M.ulcerans_Agy99 GSFEADPERVADVMSLEEKEAIRDHGLLIKCGIVLLLVFTAFVANSALHV
MMAR_2030|M.marinum_M GSFEADPERVADVMSLEEKEAIRDHGLLIKCGIVLLLVFTAFVANSALHV
MAV_3579|M.avium_104 GAFTVDPERVADVMSLEEKEAIRDPRLLVTCGVVLLAVFAAFIAHGPLHL
Mb2703|M.bovis_AF2122/97 GSITVEADRIADVMALDEGEAIRDRGLLVKCGAVLVLVFAAFVAHPVLHI
Rv2684|M.tuberculosis_H37Rv GSITVEADRIADVMALDEGEAIRDRGLLVKCGAVLVLVFAAFVAHPVLHI
MLBr_01036|M.leprae_Br4923 GAFAVDPERVADIMSLNEREAIRDRWLLIKCGVVLLLVFVAFIAHPVLHT
MSMEG_0851|M.smegmatis_MC2_155 KELRANHVHIEEVMALQERRAIKDMRLLVRSLIVLNIVIVGFALHSVFHV
Mflv_1448|M.gilvum_PYR-GCK KDLATEPQR-AVSVEP--------LDVPKFTLAVVVLTLAGFAVTSLLGF
Mvan_5336|M.vanbaalenii_PYR-1 KDLAVEPRR-AAVTEP--------VEVPVFALVVLGLTLLGFAVTSLLGL
TH_3461|M.thermoresistible__bu ADLSPAPSTGDRPAEP--------VGVPVFVLTVLGLTLTGFAVTSWLGI
: : *: .: .* :
MUL_3321|M.ulcerans_Agy99 KPSMVALIGAGILVVVSRLDHSDYLSSVEWDTLLFFVGLFIMVGALVKTG
MMAR_2030|M.marinum_M KPSMVALIGAGILVVVSRLDHSDYLSSVEWDTLLFFVGLFIMVGALVKTG
MAV_3579|M.avium_104 EPSLVALLGAGILVVASRLQPADYLSGVEWDTLLFFAGLFVMVGALVKTG
Mb2703|M.bovis_AF2122/97 QPSLVALLGAGMLIVVSGLTRSEYLSSVEWDTLLFFAGLFIMVGALVKTG
Rv2684|M.tuberculosis_H37Rv QPSLVALLGAGMLIVVSGLTRSEYLSSVEWDTLLFFAGLFIMVGALVKTG
MLBr_01036|M.leprae_Br4923 GPSLVGMLGAGILIVISKLERSDYLSSVKWETLLFFAGLFIMVGALVKTD
MSMEG_0851|M.smegmatis_MC2_155 APSIVALLGAGVMLLVTDVDINEVLPEVEWPTLVFFMGLFVMVAGLTHTG
Mflv_1448|M.gilvum_PYR-GCK SPAWAALAGALVLGVRALADRRTTMSRIVVSLDIPFLIFVLCLGVVVDAV
Mvan_5336|M.vanbaalenii_PYR-1 SPAWAALAGALVLGVRALATRHTTVTRIAAAADVPFLIFVLCLGVVVAAV
TH_3461|M.thermoresistible__bu APAWAALAGALVLGVHGLARGRTTAEGVLSAANLPFLAFVVCLGIVVEAV
*: ..: ** :: : : : * :.: :. :. :
MUL_3321|M.ulcerans_Agy99 VVTQVAQMAVTATG-GNTLLATMVILASSLVISGIVNNVPYAATMTPIVA
MMAR_2030|M.marinum_M VVTQVAQMAVTATG-GNTLLATMVILAASLVISGIVNNVPYAATMTPIVA
MAV_3579|M.avium_104 VVKHLARLAITATG-GNTLTATMVILVASVVISGIVDNVPYAATMAPVVA
Mb2703|M.bovis_AF2122/97 VVNDLARAATQLTG-GNIVATAFLILGVSAPISGIIDNIPYVATMTPLVA
Rv2684|M.tuberculosis_H37Rv VVNDLARAATQLTG-GNIVATAFLILGVSAPISGIIDNIPYVATMTPLVA
MLBr_01036|M.leprae_Br4923 VVNQLARATTTLTG-GHELLTVVLTLGVSTLVSSIIDNIPYVATMTPIVS
MSMEG_0851|M.smegmatis_MC2_155 VIGELGSVAQSAFG-DNWLGAATALLFGSSIAGAFIDNIPYTATMTPVVE
Mflv_1448|M.gilvum_PYR-GCK MRNGLESAMQQVIPDGHGLLALLGIAAVAALLSNVVNNLPAVLVLLPLVA
Mvan_5336|M.vanbaalenii_PYR-1 MRNGLESAMRDAIPEGQGLAALLGIAAVAAVLSNVVNNLPAVLVLLPLVT
TH_3461|M.thermoresistible__bu RTAGLDGALAAVLPDPGSLWGLLGIAAAAAVLANAVNNLPAVLALLPLVS
: : : . ::*:* . .: *:*
MUL_3321|M.ulcerans_Agy99 ELVPVLVDKGNPDVLWWALALGTDFGGDLTAIGASANVIVLGIAKRADNP
MMAR_2030|M.marinum_M ELVPVLVDKGNPDVLWWALALGTDFGGNLTAIGASANVIVLGIAKRADNP
MAV_3579|M.avium_104 DLVPALGDHANPAVLWWSLALGTDFGGNLTAIGASANIVLLGIARRADNP
Mb2703|M.bovis_AF2122/97 ELVAVMGGQPSTDTPWWALALGADFGGNLTAIGASANVVMLGIARRAGAP
Rv2684|M.tuberculosis_H37Rv ELVAVMGGQPSTDTPWWALALGADFGGNLTAIGASANVVMLGIARRAGAP
MLBr_01036|M.leprae_Br4923 ELVASMPDQSHTDILWWALALGADFGGNLTAVGASANVVMLEIAKSAGTP
MSMEG_0851|M.smegmatis_MC2_155 SMVADAPDMVTGNALWWAFALGACFSGNGTAIAASANVVAIGIAKRAGHP
Mflv_1448|M.gilvum_PYR-GCK GSGPAAV---------LAVLIGVNIGPNLTYVGSLANLLWRSVVR-RDVK
Mvan_5336|M.vanbaalenii_PYR-1 PIGPAAV---------LAVLIGVNIGPNLTYVGSLANLLWRSVVR-RDMR
TH_3461|M.thermoresistible__bu AAGPAAV---------LAVLIGVNIGPNLSYLGSLSNLLWRNAVQ-PDIP
. :. :*. :. : : :.: :*:: .: .
MUL_3321|M.ulcerans_Agy99 ISFWEFTRKGIVVTTMSLVLAAIYLWVRYFVLS-
MMAR_2030|M.marinum_M ISFWEFTRKGIVVTTMSLVLAAIYLWVRYFVLS-
MAV_3579|M.avium_104 ISFWEFTRKGVVVTAVSVALSALYLWLRYFVWG-
Mb2703|M.bovis_AF2122/97 ISFWEFTRKGAVVTAVSIALAAIYLWLRYFVLLH
Rv2684|M.tuberculosis_H37Rv ISFWEFTRKGAVVTAVSIALAAIYLWLRYFVLLH
MLBr_01036|M.leprae_Br4923 ISFWEFTRKGIAVTVISIALAGIYLWLRYLVMS-
MSMEG_0851|M.smegmatis_MC2_155 ISFWQFTRYGIVVTFLSTALAWVYVWLRYF----
Mflv_1448|M.gilvum_PYR-GCK AGFGEFSRIGVVTTPLVLVAAVLGLWTYVRLIGL
Mvan_5336|M.vanbaalenii_PYR-1 VGFAEFSRAGLVTAPLTLVAAVLGLWAWIQLFGL
TH_3461|M.thermoresistible__bu AGFGEFSRIGLMTAPLTLVASVLALWVSLQLFGT
.* :*:* * .: : . : : :*